Back to article: The role of hydrophobic matching on transmembrane helix packing in cells
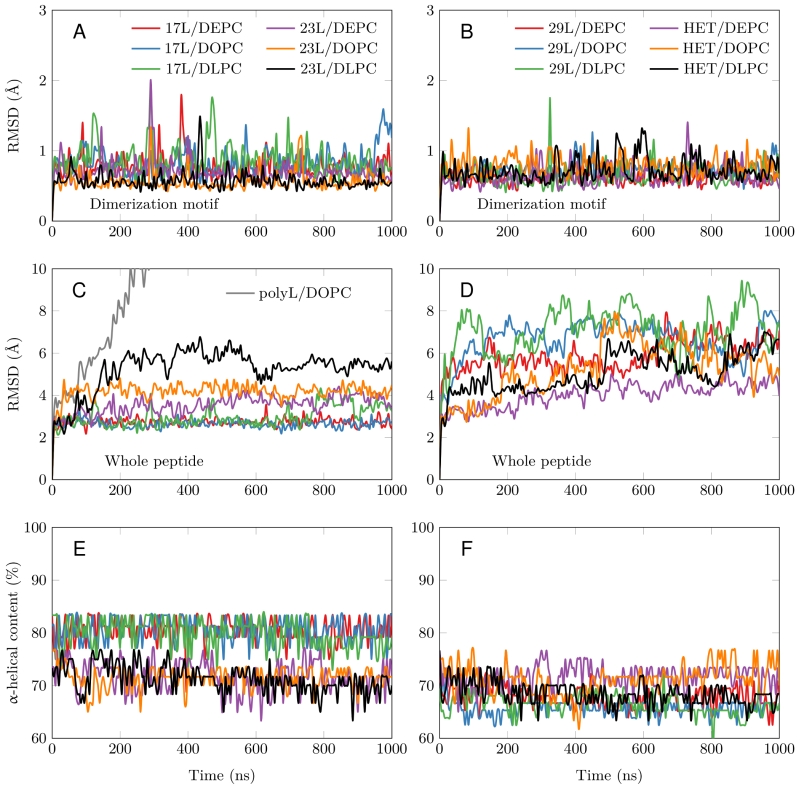
FIGURE 3: Stability of the dimeric structures from MD simulations. (A-D) Root mean squared deviation (RMSD) of the dimerization motif (5 residues per peptide) is shown on the top row and the RMSD of the whole dimer on the bottom row. The whole trajectory is included in the analyses. The 17L/29L hetero-dimer is labeled “HET”, and the polyleucine control in DOPC as “PolyL/DOPC”. (E and F) Helicity of the peptides during the full simulation time period (color code as in At to D). The 17L/29L hetero-dimer is labeled “HET”.