Reviews:
Cell Stress, Vol. 5, No. 9, pp. 128 - 142; doi: 10.15698/cst2021.09.255
Atg8ylation as a general membrane stress and remodeling response
1 Autophagy Inflammation and Metabolism Center of Biomedical Research Excellence, University of New Mexico Health Sciences Center, Albuquerque, NM 87131, USA.
2 Department of Molecular Genetics and Microbiology, University of New Mexico Health Sciences Center, Albuquerque, NM 87131, USA.
Keywords: ubiquitylation, endosome, exosomes, microvesicles, secretory autophagy, unconventional secretion, secretion, ubiquitin, galectin, lap, lc3, atg8, tfeb, lysosome, ampk, mtor, autophagy.
Abbreviatons:
AIM – Atg8-interacting motif;
EV – extracellular vesicle;
ILV – intralumenal vesicle;
LANDO – LC3-associated endocytosis;
LAP – LC3-associated phagocytosis;
LIR – LC3 interacting region;
mAtg8s – mammalian Atg8s;
mTOR – mechanistic target of rapamycin;
PAMP – pathogen associated molecular pattern;
PE – phosphatidylethanolamine;
PS – phosphatidylserine;
ROS – reactive oxygen species; SLR – sequestosome-like receptor;
TFEB – transcription factor EB;
TLR- Toll-like receptor;
UPS – ubiquitin proteasome system.
Received originally: 29/05/2021 Received in revised form: 26/07/2021
Accepted: 30/07/2021
Published: 12/08/2021
Correspondence:
Vojo Deretic, Department of Molecular Genetics and Microbiology, University of New Mexico Health Sciences Center, Albuquerque, NM 87131, USA; vderetic@salud.unm.edu
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Suresh Kumar, Jingyue Jia and Vojo Deretic (2021). Atg8ylation as a general membrane stress and remodeling re-sponse. Cell Stress 5(9): 128-142. doi: 10.15698/cst2021.09.255
Abstract
The yeast Atg8 protein and its paralogs in mammals, mammalian Atg8s (mAtg8s), have been primarily appreciated for their participation in autophagy. However, lipidated mAtg8s, including the most frequently used autophagosomal membrane marker LC3B, are found on cellular membranes other than autophagosomes. Here we put forward a hypothesis that the lipidation of mAtg8s, termed ‘Atg8ylation’, is a general membrane stress and remodeling response analogous to the role that ubiquitylation plays in tagging proteins. Ubiquitin and mAtg8s are related in sequence and structure, and the lipidation of mAtg8s occurs on its C-terminal glycine, akin to the C-terminal glycine of ubiquitin. Conceptually, we propose that mAtg8s and Atg8ylation are to membranes what ubiquitin and ubiquitylation are to proteins, and that, like ubiquitylation, Atg8ylation has a multitude of downstream effector outputs, one of which is autophagy.
INTRODUCTION
The yeast Atg8 protein and its seven mammalian Atg8 paralogs (mAtg8s) that include LC3A, LC3B, LC3B2, LC3C, GABARAP, GABARAPL1, and GABARAPL2/GATE16 [1][2][3] are best known for their role in autophagy [4], a process described early on along with the definition of lysosomes [5][6]. Canonical autophagy is a metabolic and cellular quality control process [7] which typically sequesters cytoplasmic cargo into double-membrane autophagosomes decorated with mAtg8s [8] and typically delivers the cargo to autolysosomes via fusion between autophagosomes and lysosomes [9]. The default termination of the canonical pathway in autolysosomes is degradation of the captured cytoplasmic material [8]. However, mAtg8s are found on a variety of other membranes in diverse biological and physiological contexts, including LC3-associated phagocytosis (LAP) and its variations [10]. Currently, many of these and related phenomena are grouped under the umbrella of ‘non-canonical autophagy' [10], although some of them such as LAP lack cytosolic cargo, which in principle defines the term ‘autophagy' or ‘self-eating'. As an alternative to ‘non-canonical autophagy', here we propose the ‘Atg8ylation hypothesis' as a principle that can unify different manifestations and roles of mAtg8s, their lipidation, and their association with various membranes. We propose that mAtg8ylation is a cellular response that both counters membrane stress and is a mechanism involved in general membrane remodeling, with canonical autophagy being one manifestation.
–
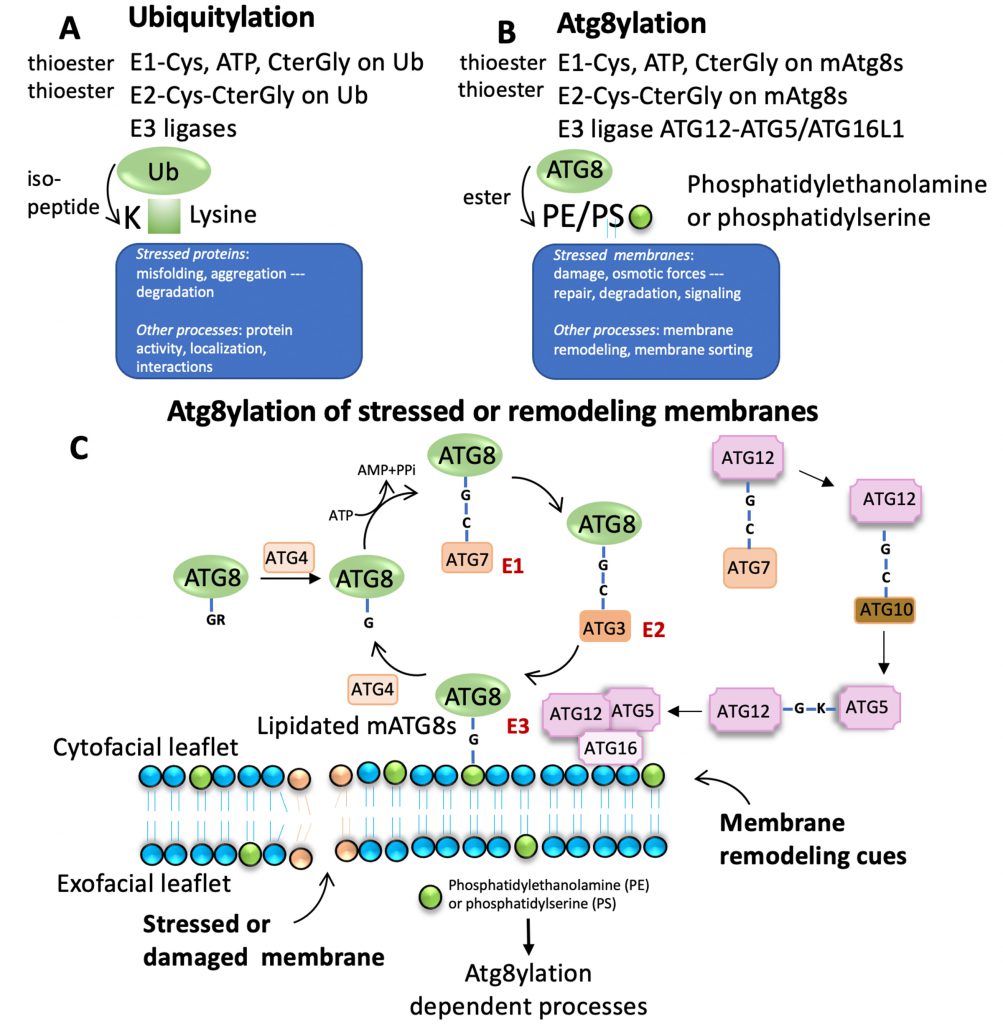
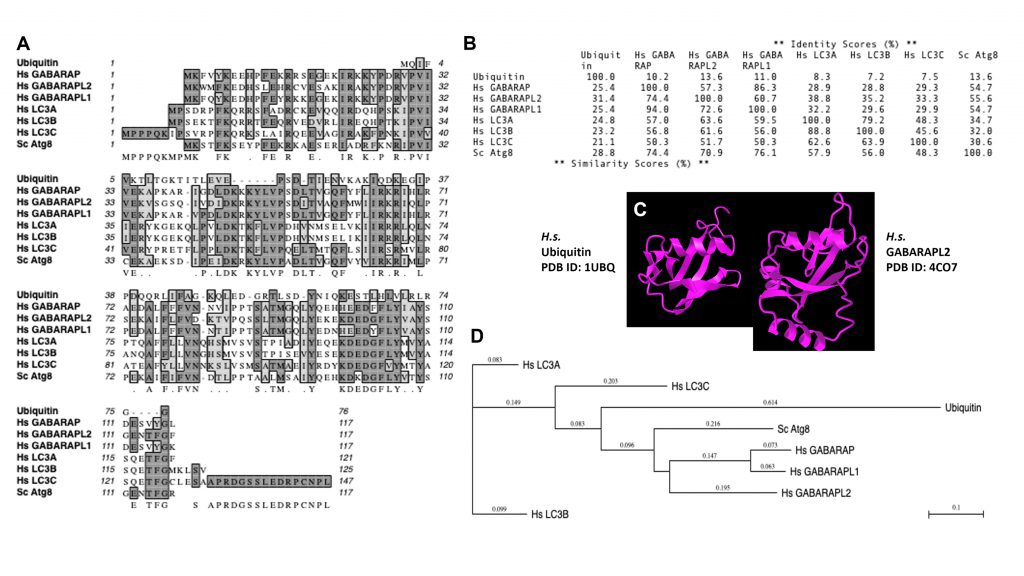
Of note, mAtg8s are related to ubiquitin in sequence and structure (Figures 1 and 2). Ubiquitylation is associated with disassembly, degradation and removal of misfolded proteins triggered by stressors [11][12][13]. Ubiquitin protein modifications [14] also play other non-degradative roles, such as modulation of normal protein activity, localization and interactions under unperturbed conditions [11][12][13]. We hypothesize that mAtg8s and Atg8ylation play a role in processes maintaining membrane homeostasis under stress conditions as well as in normal membrane remodeling in response to programmed, physiological or pathological cues. We propose here that Atg8ylation (Figure 1) is to membranes what ubiquitylation is to proteins.
–
ATG8YLATION AND UBIQUITYLATION
Both autophagy [8], introduced above, and the ubiquitin-proteasomal system (UPS) [11][12] are major modulatory machineries in eukaryotic cells which are often engaged in cargo degradation but are also performing multiple other functions [10][13][15]. Both systems require tagging of the cargo/target to be processed or acted upon.
–
Ubiquitylation of proteins often occurs on misfolded and aggregated proteins caused by stressors as well as under unperturbed conditions, modulating normal protein activity, localization, and their interactions (Figure 1A). Atg8ylation is a process whereby the Atg8 conjugation machinery is recruited to damaged or otherwise stressed membranes or to membranes undergoing remodeling under various homeostatic or non-homeostatic conditions (Figure 1B) and catalyzes mAtg8s' conjugation to phosphatidylethanolamine (PE) [16][17][18], or phosphatidylserine (PS) [18]. Like ubiquitylation, Atg8ylation involves an E1-like activating protein, ATG7 and requires ATP for activation (Figure 1C). ATG7 conjugates to the C-terminal glycine of mAtg8s, exposed post-translationally by ATG4 proteases. This is followed by action of the E2-like activity of ATG3 on ATG8s. Finally, the conjugation of mATG8s to PE or PS is mediated by an E3-like complex ATG5-ATG12/ATG16L1 (Figure 1C) [4][19].
–
Of note, the diverse mammalian Atg8s are all related to ubiquitin (Figure 2A-C), underscoring the similarities between the two systems and the biochemical relatedness of ubiquitylation and Atg8ylation consistent with the parallels in the conjugation cascade. The two differ principally in the type of targets that ubiquitin or Atg8s are being conjugated to – in the case of ubiquitin to the stressed and degradation-bound or otherwise modified proteins, whereas in the case of mAtg8s, to the stressed or otherwise engaged membranes. The diverse events downstream or associated with Atg8ylation are individually described in subsections below.
–
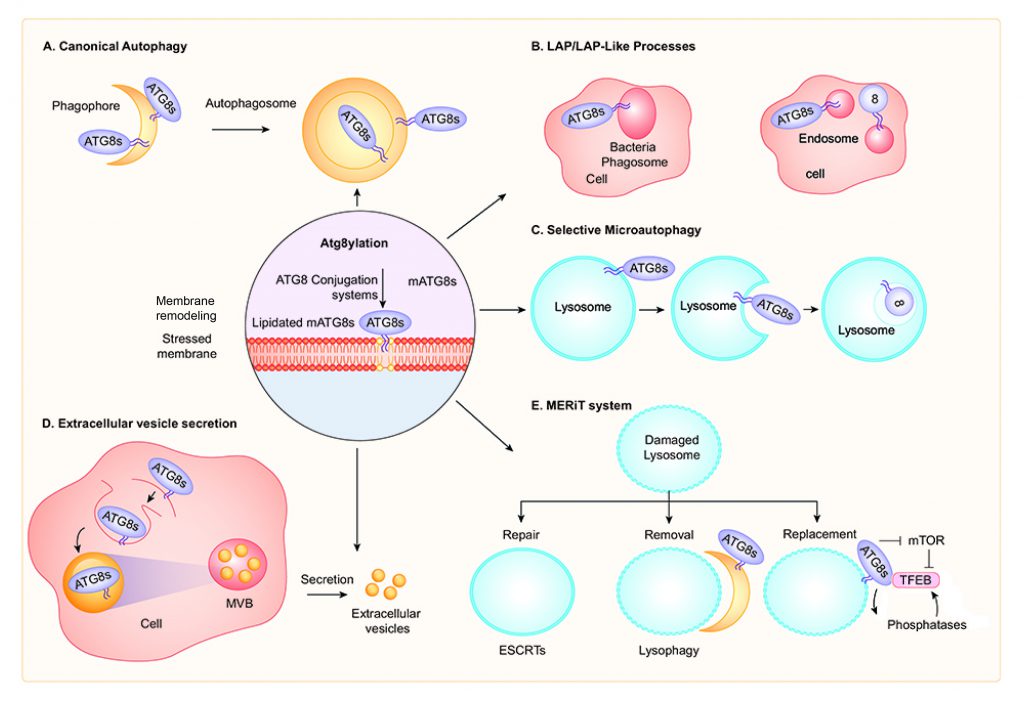
CANONICAL AUTOPHAGY
As mentioned above, mAtg8s and Atg8ylation have been initially almost exclusively associated with the process of autophagy [5][6] controlled by an ensemble of Atg factors, first genetically defined in yeast [12][20][21] and conserved in mammals [8][19]. Canonical autophagy has been appreciated primarily as a degradative process [4], with metabolic and quality control functions [7]. Whereas the degradative function of UPS involves elimination of individual proteins [22], autophagy can eliminate protein aggregates and larger structures, including whole organelles or their parts [23]. Autophagy is initiated in response to various cellular stressors, and requires the lipid kinase VPS34 (PIK3C3), first identified in yeast as a PI3P-generating enzyme in response to stress such as membrane-stretching osmotic changes [24]. The details of the autophagy pathway have been reveled through genetic analyses in yeast [4][12][20][21] with a suite of ATG genes controlled by upstream kinases AMPK [25][26][27], mTOR (mechanistic target of rapamycin) [26][28][29] and TBK1 [30][31][32][33][34].
–
Conventionally, autophagy initiation is controlled by several modules [19]: (i) The ULK1/2 kinase complex with FIP200, ATG13 and ATG101, acting as conduits for inhibition by active mTOR [28][29][35] and activation by AMPK [25] to induce autophagy; (ii) ATG14L-endowed Class III PI3-Kinase Complex [36][37][38] that includes VPS34 and Beclin 1 [39], which can also be modified by AMPK to specifically activate the ATG14L form of VPS34 [40]; (iii) ATG9, and the ATG2-WIPI protein complexes [41][42][43]. These modules become interconnected, via FIP200 that bridges the ULK1/2 complex with the mAtg8s conjugation system by binding ATG16L1 [44][45][46], via ATG16L1 and WIPI interactions [47], and ATG13 connecting the ULK1/2 complex with ATG14-VPS34 [48][49]. After initiation, autophagy terminates in a merger of autophagosomes with degradative endolysosomal compartments whereby the sequestered cargo is degraded. This is catalyzed by a suite of SNAREs, e.g. STX17 [50] YKT6 [51], VAMP8, and SNAP29 [50].
–
Canonical autophagy and its variations manifested in different forms of selective or quality control autophagy, defined by the captured cargo (mitophagy, ER-phagy, aggrephagy, xenophagy, ribophagy, pexophagy, lipophagy, and precision autophagy of individual protein complexes) [7][8] depend on the above apparatus to be guided by the receptors and receptor-regulators, many of whom belong to the category termed Sequestosome-like receptors (SLRs) named after the founding member Sequestosome 1 (SQSTM1/p62) [52]. The majority of autophagic receptors interact with mAtg8s, albeit interaction directly with FIP200 is another “entry” into the pathway [33][34][53][54], likely amplified by eventual mAtg8 binding [53]. To interact with mAtg8s, SLRs contain dedicated Atg8 interacting motifs (AIM), also known as LIRs (LC3 interacting regions). As an interesting crossover to the ubiquitin system, SLRs often recognize ubiquitylated cargo via dedicated ubiquitin binding domains [55]. Thus, Atg8ylation and ubiquitylation act here as coincidence detectors for efficient cargo degradation.
–
Autophagosomal membrane formation is believed to entail mAtg8 conjugation machinery and Atg8ylation, i.e. Atg8 lipidation on membranes that form autophagosomes (Figure 3A) [19]. However, recent studies indicate that mAtg8s may not be absolutely required to form autophagosomal membranes as their absence mostly have limited kinetic or autophagic vesicle size effects [56][57]. This is despite mAtg8's iconic status, since LC3B is used as an almost quintessential marker of autophagosomes [58]. Atg8ylation may play additional roles in autophagy, for example as a modulator of the recruitment and/or function of SNAREs. This includes a large subset of AIM (LIR)-containing SNAREs including STX3, STX4, STX6, STX16, STX17, STX19, Vti1a, GOSR1, and VAMP7 [59][60][61], several of which are important in control of general membrane fusion and flow, as well as specifically for autophagosomal [32], autolysosome [50], and lysosomal [61] biogenesis. There are indications that Atg8ylation plays a role in modulating these SNARE-controlling mAtg8 activities, since expression of a dominant negative ATG4 that prevents lipidation of all mAtg8s inhibited STX17 function [60] in the autophagosomal pathway.
LAP AND LAP-RELATED PROCESSES
LAP [10][62], LANDO (LC3-associated endocytosis) [63], and a cluster of other related phenomena involving phagosomes or stressed endosomes [64][65][66][67][68] to which we refer collectively here as Lapoid processes, represent a form of Atg8ylation occurring on single membranes of endosomes and phagosomes (Figure 3B). LAP per se lacks cytosolic cargo, but some Lapoid processes involve cytosolic cargo. LAP involves Atg8ylation, specifically of LC3 subfamily of mAtg8s [69], on membranes of phagosomes taking up stress- or signal-inducing cargo such as potentially dangerous pathogens, inflammation-inducing dead cells, live cells to be eliminated by entosis, and extracellular debris or aggregates [62][65][68][69][70]. Of note, not every endosome and phagosome undergoes Atg8ylation, and it takes a specific stress or danger signal such as presence of TLR (Toll-like receptor) ligands within the phagocytosed or endocytosed material [62]. TLR signaling recognizes fungal, bacterial and microbial products commonly known as PAMPs (pathogen associated molecular patterns), and in principle induces strong Atg8ylation of endomembranes [71].
–
–
The key distinguishing feature of LAP, LANDO, and other Lapoid processes that differentiates them from canonical autophagy is the absence of double membranes, i.e., Atg8ylation occurs on the delimiting single bilayer membrane of the stressed endosomes and phagosomes. Further key differences are the dependence of LAP on RUBCN (Rubicon), which is an inhibitor of canonical autophagy, and its independence of FIP200, which is required for canonical autophagy and Atg8ylation of conventional double membrane autophagosomes.
–
In principle, a similar mAtg8 lipidation machinery is involved in Lapoid phenomena as in the case of canonical autophagy. It is the ATG16L1 component of the Atg8ylation E3 ligase (ATG5-ATG12/ATG16L1) that discerns between target membranes that are to be Atg8ylated [17]. Lipid binding domains are found in ATG16L1 that govern canonical autophagy vs. Lapoid phenomena [17]. The C-terminal membrane-binding region of ATG16L1 is dispensable for canonical autophagy but important for Lapoid processes [17]. The N-terminal membrane-binding amphipathic helix of ATG16L1 is required for Atg8ylation (specifically demonstrated in LC3B lipidation assays) in both canonical autophagy and Lapoid phenomena [17].
–
The upstream machinery that recognizes stressed membranes remains to be elucidated, but may involve signals form TLRs, stretching of lipid bilayers during osmotic stress, ion transport events, etc. One upstream signal that has been clearly established in LAP is NOX2 (NADPH oxidase) [72], which is classically activated upon opsonized-pathogen uptake by phagocytic cells to generate reactive oxygen species (ROS) [73]. Whereas it is not completely clear how NOX2 or NOX-generated ROS function to promote LAP, ROS can inhibit ATG4, an enzyme catalyzing mAtg8 dilapidation reaction (equivalent to DUB activity on ubiquitinated proteins) [74]. This could locally, i.e., in the vicinity of a phagosome undergoing LAP, stabilize lipidated mAtg8s.
–
How do LAP or Lapoid processes terminate, is another open question. Phagosomes that are Atg8ylated (LAP) appear to fuse with lysosomes in a more efficient way [62]. The mechanism of how mAtg8s increase fusion of LAPosomes may be similar to that of how autophagosomes fuse with lysosomes. It is possible that Atg8ylation in the case of LAP stimulates an equivalent process as in the case of Atg8ylation recruiting/activating STX17 on autophagosomes via AIM/LIR motifs [60].
–
LAP and Lapoid functions include anti-inflammatory processes such as those failing in lupus [75], elimination of pathogens [72][76], removal of dead cells and efferocytosis [77][78], entosis [65], and removal of rod outer segments via phagocytosis by retinal pigment epithelium cells [79]. LAP may promote cancer progression by favoring immune-tolerance in the cancer microenvironment, whereas defects in LAP elicit pro-inflammatory cytokines in tumor-associated macrophages [80]. LAP, however, can be pro-inflammatory by promoting TLR signaling through IRF7, which leads to type I interferon response [81]. LANDO deficient microglia display hyper-neuroinflammation and neurodegeneration; LANDO may be important for clearance of β-amyloid aggregates and suppression of microglia activation [63]. Physiological functions of many other Lapoid processes are yet to be defined.
SELECTIVE MICROAUTOPHAGY
Selective microautophagy of mammalian lysosomal membranes (Figure 3C) and its proteins occurs in response to osmotic stress or glucose-starvation, adjusting the size of lysosomes under stress conditions [82]. This requires lipidation of mAtg8 and indicates that Atg8ylation participates in microautophagy [82]. It is unaffected by inactivation of ATG13, a member of the ULK1-FIP200-ATG13-ATG101 complex, akin to other Lapoid activities. This is another example of the mATG8s lipidation machinery being recruited to single bilayer endolysosomal membranes and serves here specifically to downsize lysosomes or control their surface-to-volume characteristics under stress conditions.
–
It is unclear how other microautophagy processes differ from the selective microautophagy described above. In principle, microautophagy represents a collection of processes, best characterized in yeast, which involve direct internalization of cytoplasmic cargo into the lumen of lysosomes by invaginations of the delimiting single bilayer lysosomal membrane [83][84][85][86]. Yeast microautophagy requires the action of most Atg proteins [86], and may be different from microautophagy (endosomal, multivesicular bodies, and selective microautophagy) in mammalian cells. The selective microautophagy phenomena in principle engage components of the ATG conjugation machinery [82], ESCRT machinery [87][88], and a subset (but not all) SLRs [88], and may in some instances depend on Atg8ylation. Starvation-induced degradation of SLRs, other selective autophagy receptors, and ferritinophagy receptor NCO4 is independent of mTOR, which negatively controls canonical autophagy, and, consistently with that, depletion of FIP200, an essential component of canonical autophagy that transduces mTOR signals [89], does not prevent starvation-induced degradation of certain SLRs and NCAO4 [88]. Nevertheless, depletion of ATG7 blocks (at least in part) degradation of certain SLRs, specifically p62 and NDP52 [88], this strongly suggests that this process likely involves Atg8ylation on respective endolysosomal membranes. It is not known whether and to what extent Atg8ylation affects classical mammalian endosomal microautophagy, e.g. multivesicular bodies.
–
In another example of the role of Atg8ylation in selective microautophagy, excess ER during recovery from ER stress undergoes piecemeal micro-ER-phagy and requires mAtg8 lipidation [87].
UNCONVENTIONAL SECRETION: EXTRACELLULAR VESICLES AND SECRETORY AUTOPHAGY
The emerging role of Atg8ylation in generating extracellular vesicles (EVs) mirrors its role in selective microautophagy. Microautophagy directly carried by lysosomes and endosomes can in principle result in sequestration, degradation or secretion of intralumenal vesicles (ILVs). The exocytosed ILVs in the extracellular space are referred to as exosomes, a subtype of EVs [90]. EVs are a collection of heterogeneous membranous structures coming in variety of forms and ranging from submicron to several microns in size and include microvesicles (also known as ectosomes) and exosomes [91]. Microvesicles directly bud off the plasma membrane after its outwardly evagination, whereas exosomes originate from endomembranous ILVs, both, however, undergoing the same, exofacial direction of budding [92]. EVs are present in biological fluids with a multitude of physiological effects [92], but are yet to be fully defined in terms of specificity of cargo packaging and associated functions [93]. EVs were originally identified as vehicles for selective elimination of proteins, lipids and RNA from cells [92][94], however, EVs have now emerged as a means of intercellular communication [92][95][96].
–
Atg8ylation participates in the formation of exosomes [97][98] and secretion of specific cytosolic cargo by EVs [99] (Figure 3D). Proteomic and RNA profiling of EVs identified diverse RNA binding proteins and small non-coding RNAs requiring LC3 and the Atg8ylation machinery for packaging and secretion [99]. Among other potential functions reported for exosomes impacted by Atg8ylation, one is the decoy sequestration of bacterial membrane damaging toxins before they can reach and attack the cellular plasma membrane [100].
–
Atg8ylation also plays a role in the type of unconventional secretion or excretion/extrusion of cytoplasmic material referred to as secretory autophagy [101][102], with cargo ranging from individual proteins to intracellular organelles or pathogenic microorganisms [101], such as mycobacteria [103] and specific viruses [104]. Atg8ylation furthermore promotes certain types of viral budding at the plasma membrane, as in the case of filamentous mode of budding of influenza A [105]. The prototypical cytokine cargo for secretory autophagy are IL-1β [106][107][108][109], which does not have a leader peptide and thus is synthesized as a cytosolic protein [110], and additional cytokines such as IL-6 [111] and alarmins such as HMGB1 [107][112]. However, IL-1β release from cells utilizes multiple routes. Such alternative pathways include import of the cytosolic IL-1β into the lumen of ERGIC, an intermediate compartment within the canonical secretory pathway [113][114]. The most dominant route of passive release of IL-1β from dying cells is through gasdermin pores on the plasma membrane during pyroptosis [115], and IL-1β leaks through such pores even before the ‘official' cell death [116]. Whether and how Atg8ylation interfaces with various forms of unconventional secretion [117] remains to be fully explored and associated mechanisms understood.
ATG8YLATION AS A RESPONSE TO MEMBRANE STRESS AND DAMAGE
Biological membranes provide a key diffusional barrier and define the physical boundaries of cells and their intracellular compartments; they are endowed with selective permeabilities, channels, pumps, and signaling systems [118][119]. A number of stress conditions affect integrity and function of cellular membranes [120], which are in principle two-dimensional liquids of hydrated lipid bilayers composed of phospholipids, sterols, sphingolipids, integral and peripheral membrane proteins often decorated by glycocalyx [119]. To counter inherent membrane fragility, all cells have mechanisms to counter and repair damage [120], with morphologically visible measures taken by microbes, fungi and plants that stabilize their cells by rigid cell walls countering the osmotic stress among other environmental insults. Physical membrane damage [120], programmed or unprogrammed permeabilization transitions [115][121][122], osmotic stress [123], lipid tension or fluidity changes due to composition or temperature changes, exposure to microbial and environmental toxins and detergent-like molecules, as well as entropic changes, if not countered by continuous repair/replenishment functions [120], can perturb membranes either functionally or physically, with manifestations in pathological states and disease [118].
–
The plasma membrane is by its boundary nature under continuous risk of being damaged by exogenous agents, and conversely is used for programmed permeabilization events associated with cell death pathways. Recent studies [124][125][126] suggest that Atg8ylation-dependent processes and other ATG proteins engaged in non-canonical functions contribute to the protection of plasma membrane against physical damage (modeled by laser damage or detergent treatment), programmed permeabilization by gasdermin-pores during pyroptosis or MLKL during necroptosis, by MLKL-like permeabilization function of SARS-CoV-2 ORF3a, bacterial pore forming toxins (pneumolysin, lysteriolysin and streptolysin O), and harsh components of the cell walls of microbes such as Mycobacterium tuberculosis[124][126]. Whereas ATG9A engages ESCRT proteins to seal plasma membrane holes immediately upon permeabilization [126], Atg8ylation results in blebbing due to lysosomal exocytosis [124] or endocytic processes termed LC3-associated micropinocytosis which appears to be a Lapoid process [125].
–
Intracellular membranes can also come under the physical assault or be subjected to membrane stress, such as osmotic changes. The lysosome is the digestive organelle of the cell and thus receives cargo destined for degradation from both cellular interior and the exterior space, via autophagic, endosomal and phagocytic pathways [127]. By the very nature of receiving various material from different tributary pathways, lysosomal membranes and organelles of the endolysosomal system are at risk of being damaged by the ingested material [128], lysosomotropic compounds [129], osmolytes such as trehalose [130], neurotoxic protein aggregates including α-synuclein, tau, and huntingtin amyloids [131][132][133], mineral crystals such as silica, monosodium urate, and calcium phosphate [134][135][136], pathogens, PAMPs and DAMPs (danger associated molecular patterns) [137][138], and can be influenced by programmed events such as TRAIL (tumor necrosis factor-related apoptosis-inducing ligand) and downstream signaling [122][131][139][140][141][142].
–
Lysosomal damage elicits a set of diverse responses to restore lysosomal homeostasis [132][143][144][145][146][147][148], reinstate key functions including delivery of iron [149], and adjust cellular metabolic needs by suppressing mTOR and stimulating AMPK while simultaneously mobilizing homeostatic responses requiring mTOR inhibition and AMPK activation [142][150]. Early dominant events include responses by galectins [133][138][143][151][152], ubiquitin [132][137][142][144][145], ESCRTs [133][146][147], and include additional events [148] including recruitment of mAtg8s [153], which engage not only in canonical autophagy [46][135] but also in other Atg8ylation-dependent processes [154][155] elaborated in the subsequent section. Eventually, if membrane repair fails, a type of canonical autophagy termed lysophagy [46][133][135][143][145][150] eliminates excessively damaged lysosomes, endosomes or phagosomes [138].
ATG8YLATION SIGNALING AND TFEB RESPONSE IN LYSOSOMAL BIOGENESIS
Atg8ylation contributes to signaling events that activate TFEB (transcription factor EB) to initiate the lysosomal protein expression program. Starvation and additional signals, e.g., lysosomal damage, induce TFEB translocation form the cytosol to the nucleus where it stimulates lysosomal gene expression, and to an extent several autophagy genes [156][157][158]. Apart from its role in lysosomal biogenesis, TFEB transcriptionally controls other biological processes [127] including lipid catabolism [159], lysosomal exocytosis [160], mitochondrial biogenesis [161] and mitophagy [162]. TFEB activity is controlled by mTOR [127][163][164]. TFEB phosphorylation by mTOR increases its binding to 14-3-3 proteins, which holds TFEB in the cytosol [163][165]. TFEB is under a special control by mTOR, downstream of the inputs from Foliculin/FLCN to RagC/D, in contrast to other signals transduced to mTOR via RagA/B [166]. Protein phosphatase 3 catalytic subunit beta (PPP3CB) dephosphorylates TFEB and releases it from 14-3-3 [165]. Additional studies reporting calcium-dependent but PPP3CB-independent dephosphorylation of TFEB indicate possible involvement of another phosphatase [155].
–
TFEB is activated by a variety of membrane and other stress conditions including starvation [154][164], lysosomal damage [133][143][155], pathogen infections [154][167][168][169][170] and mitochondrial stress [162]. Atg8ylation controls nuclear translocation of TFEB [154][155][171] (Figure 3E). TFEB interacts directly with GABARAP [154] and the GABARAP subset of mAtg8s supports nuclear translocation and transcriptional activity of TFEB during starvation [154] and lysosomal damage [155]. Calcium flux stimulates nuclear translocation of TFEB [155][165]. Atg8ylation affects intracellular calcium levels [154], whereas transcription of the lysosomal calcium channel TRPML1 [165] is reduced in mAtg8 depleted cells [154]. Furthermore, TRPML1 interacts with lipidated LC3 in response to lysosomal damage [155], which is a clear example of Atg8ylation controlling molecular events other than lysophagy.
–
Atg8ylation has appreciable effects on TFEB activity during starvation, lysosomal damage of xenophagy [154][162], [155][171]. GABARAP binds FLCN/FNIP complex [171], which is a GAP (GTPase activating protein) for RagC/D GTPase, and activates (paradoxically) mTOR when amino acids are aplenty [172][173]. However, under membrane stress conditions, binding of FLCN to membranes Atg8ylated by GABARAP disrupts the regulation of RagC/D by FLCN, impairing TFEB phosphorylation by mTOR and activating TFEB [171]. In conclusion, Atg8ylation has significant and multiple roles to in control of TFEB activity.
CODA
The herein proposed unifying concept of Atg8ylation refers to a process of tagging stressed membranes with lipidated mAtg8s. This includes damaged, or otherwise mobilized and remodeling membranes leading to a plethora of outcomes. Often, these processes occur on endosomal, lysosomal and autophagosomal membranes but may not be limited to these organelles. The spectrum of presentations includes canonical autophagy, LAP and Lapoid processes, selective microautophagy and unconventional secretion, different stages of the MERiT lysosomal QC system, and membrane modification during programmed or signaling events.
–
The elucidation of the canonical autophagy pathway in yeast has provided a clear linear pathway of Atg8-driven steps, from autophagosome formation to their fusion with the vacuole/lysosome. In mammalian systems, this pathway and its core components are conserved. The presence of multiple Atg8 paralogs in mammals reflects increased complexity, whereas experimental analyses have revealed that mAtg8s function in processes other than autophagy. The proliferation of these ‘non-canonical autophagy-related' processes may at first appear confusing. The Atg8ylation concept proposed here resets the view of what lipidation of mAtg8s represents considering its standalone manifestation and nature – it is a covalent modification of membranes the way ubiquitin covalently modifies proteins. Once this point of view is considered, it is relatively easy to understand how this can participate in a multitude of processes, of which canonical autophagy is just a subset. This could be useful as more and more ‘anomalies' or non-canonical processes are being described, and should spur further investigations based on the principle of Atg8tylation freed from the conceptual confines, however elegant, of the canonical autophagy pathway. Retrospectively, membrane processes such as LAP, LANDO, selective microautophagy, EV/exosome secretion, etc., can be easier to understand from this point of view, while stimulating future systematic discoveries.
–
The crossovers between ubiquitylation and Atg8ylation exist, as glimpsed from emerging examples of ubiquitylation being able to occur on complex glycolipids such as bacterial LPS (lipopolysaccharide) when introduced into the mammalian cell [174], potentially blurring the borders of specialization between ubiquitylation and Atg8ylation relative to proteins vs. membranes. Conversely, two recent reports of protein Atg8ylation [175][176] confirm that Atg8ylation and ubiquitylation are more similar than previously appreciated, although the biological significance of protein Atg8ylation is yet to be explored. In addition to proposing the concept of Atg8ylation, as the membrane modification equivalent to the ubiquitylation of proteins, this review collates a number of seemingly disparate phenomena into a unified model. The concept of Atg8ylation offers a new perspective on the system usually associated with autophagy and may help in developing approaches to pharmacologically manipulate Atg8ylation rather than the entire canonical autophagy pathway.
–
With the introduction of Atg8ylation as a concept some old questions remain, and new questions arise: What exactly are the functional and mechanistic consequences of Atg8ylation in general and in specific examples covered here and how and does the lipidation of mAtg8s affect their functionalities? Do Atg8ylated membranes share specific types of lipid modifications, changes in ordering/tension within the bilayer, or other molecular features that are specific to ‘stress' or a ‘stress' signal? When a target protein reaches a threshold of ubiquitin modifications this often leads to its proteasomal degradation; could Atg8ylation similarly operate in the context of membrane remodeling and degradation? Doe Atg8ylation also take place under basal conditions, and, if so, would there be quantitative and qualitative differences between basal vs. ‘stress' conditions? With Atg8ylation expanding the functional roles of mAtg8s to outside of the cells through unconventional secretion, do these processes serve to excrete material from the cells or take active signaling roles such as in the case of EVs in intercellular communication? What are the specific roles of mAtg8s and Atg8ylation in processes of unconventional/secretory autophagy, and do they play a role in cargo selection or vesicle formation or both?
–
As the most celebrated Atg8ylation process, canonical autophagy impacts a spectrum of pathophysiological conditions including cancer, neurodegenerative diseases, metabolic and cardiovascular disorders, autoimmunity, inflammation and infection [4][177][178]. LAP and Lapoid processes are also physiologically important [10], whereas other Atg8ylation processes are slowly establishing biological and potential medical relevance. The broader view presented here of membrane-specific (albeit not exclusive) modification via Atg8ylation should open both fundamental and translational approaches different than those based on autophagy alone.
REFERENCES
- Xin Y, Yu L, Chen Z, Zheng L, Fu Q, Jiang J, Zhang P, Gong R, Zhao S (2001). Cloning, expression patterns, and chromosome localization of three human and two mouse homologues of GABA(A) receptor-associated protein. Genomics 74(3): 408-413. 10.1006/geno.2001.6555
- He H, Dang Y, Dai F, Guo Z, Wu J, She X, Pei Y, Chen Y, Ling W, Wu C, Zhao S, Liu JO, Yu L (2003). Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J Biol Chem 278(31): 29278-29287. 10.1074/jbc.M303800200
- Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z (2010). LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. Embo J 29(11): 1792-1802. 10.1038/emboj.2010.74
- Mizushima N (2020). The ATG conjugation systems in autophagy. Curr Opin Cell Biol 63: 1-10. 10.1016/j.ceb.2019.12.001
- Deter RL, De Duve C (1967). Influence of glucagon, an inducer of cellular autophagy, on some physical properties of rat liver lysosomes. J Cell Biol 33(2): 437-449. 10.1083/jcb.33.2.437
- Deter RL, Baudhuin P, De Duve C (1967). Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol 35(2): C11-16. 10.1083/jcb.35.2.c11
- Deretic V, Kroemer G (2021). Autophagy in metabolism and quality control: opposing, complementary or interlinked functions? Autophagy 1-10. 10.1080/15548627.2021.1933742
- Morishita H, Mizushima N (2019). Diverse Cellular Roles of Autophagy. Annu Rev Cell Dev Biol 35: 453-475. 10.1146/annurev-cellbio-100818-125300
- Zhao YG, Zhang H (2019). Autophagosome maturation: An epic journey from the ER to lysosomes. J Cell Biol 218(3): 757-770. 10.1083/jcb.201810099
- Galluzzi L, Green DR (2019). Autophagy-Independent Functions of the Autophagy Machinery. Cell 177(7): 1682-1699. 10.1016/j.cell.2019.05.026
- Hershko A, Ciechanover A (1998). The ubiquitin system. Annu Rev Biochem 67: 425-479. 10.1146/annurev.biochem.67.1.425
- Tsukada M, Ohsumi Y (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333(1-2): 169-174. 10.1016/0014-5793(93)80398-e
- Mevissen TET, Komander D (2017). Mechanisms of Deubiquitinase Specificity and Regulation. Annu Rev Biochem 86: 159-192. 10.1146/annurev-biochem-061516-044916
- Swatek KN, Komander D (2016). Ubiquitin modifications. Cell Res 26(4): 399-422. 10.1038/cr.2016.39
- Zheng N, Shabek N (2017). Ubiquitin Ligases: Structure, Function, and Regulation. Annu Rev Biochem 86: 129-157. 10.1146/annurev-biochem-060815-014922
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y (2000). A ubiquitin-like system mediates protein lipidation. Nature 408(6811): 488-492. 10.1038/35044114
- Lystad AH, Carlsson SR, de la Ballina LR, Kauffman KJ, Nag S, Yoshimori T, Melia TJ, Simonsen A (2019). Distinct functions of ATG16L1 isoforms in membrane binding and LC3B lipidation in autophagy-related processes. Nat Cell Biol 21(3): 372-383. 10.1038/s41556-019-0274-9
- Durgan J, Lystad AH, Sloan K, Carlsson SR, Wilson MI, Marcassa E, Ulferts R, Webster J, Lopez-Clavijo AF, Wakelam MJ, Beale R, Simonsen A, Oxley D, Florey O (2021). Non-canonical autophagy drives alternative ATG8 conjugation to phosphatidylserine. Mol Cell 81(9): 2031-2040 e2038. 10.1016/j.molcel.2021.03.020
- Mizushima N, Yoshimori T, Ohsumi Y (2011). The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27: 107-132. 10.1146/annurev-cellbio-092910-154005
- Klionsky DJ, Banta LM, Emr SD (1988). Intracellular sorting and processing of a yeast vacuolar hydrolase: proteinase A propeptide contains vacuolar targeting information. Mol Cell Biol 8(5): 2105-2116. 10.1128/mcb.8.5.2105
- Harding TM, Morano KA, Scott SV, Klionsky DJ (1995). Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol 131(3): 591-602. 10.1083/jcb.131.3.591
- Glickman MH, Ciechanover A (2002). The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82(2): 373-428. 10.1152/physrev.00027.2001
- Johansen T, Lamark T (2011). Selective autophagy mediated by autophagic adapter proteins. Autophagy 7(3): 279-296. 10.4161/auto.7.3.14487
- Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Michell RH (1997). Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature 390(6656): 187-192. 10.1038/36613
- Kim J, Kundu M, Viollet B, Guan KL (2011). AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13(2): 132-141. 10.1038/ncb2152
- Inoki K, Kim J, Guan KL (2012). AMPK and mTOR in cellular energy homeostasis and drug targets. Annu Rev Pharmacol Toxicol 52: 381-400. 10.1146/annurev-pharmtox-010611-134537
- Alers S, Loffler AS, Wesselborg S, Stork B (2012). Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol 32(1): 2-11. 10.1128/MCB.06159-11
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH (2009). ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 20(7): 1992-2003. 10.1091/mbc.E08-12-1249
- Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X (2009). ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 284(18): 12297-12305. 10.1074/jbc.M900573200
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dotsch V, Bumann D, Dikic I (2011). Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333(6039): 228-233. 10.1126/science.1205405
- Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, Dupont N, Ornatowski W, Jiang S, Bradfute SB, Bruun JA, Hansen TE, Johansen T, Deretic V (2012). TBK-1 Promotes Autophagy-Mediated Antimicrobial Defense by Controlling Autophagosome Maturation. Immunity 37(2): 223-234. 10.1016/j.immuni.2012.04.015
- Kumar S, Gu Y, Abudu YP, Bruun JA, Jain A, Farzam F, Mudd M, Anonsen JH, Rusten TE, Kasof G, Ktistakis N, Lidke KA, Johansen T, Deretic V (2019). Phosphorylation of Syntaxin 17 by TBK1 Controls Autophagy Initiation. Dev Cell. 10.1016/j.devcel.2019.01.027
- Vargas JNS, Wang C, Bunker E, Hao L, Maric D, Schiavo G, Randow F, Youle RJ (2019). Spatiotemporal Control of ULK1 Activation by NDP52 and TBK1 during Selective Autophagy. Mol Cell 74(2): 347-362 e346. 10.1016/j.molcel.2019.02.010
- Ravenhill BJ, Boyle KB, von Muhlinen N, Ellison CJ, Masson GR, Otten EG, Foeglein A, Williams R, Randow F (2019). The Cargo Receptor NDP52 Initiates Selective Autophagy by Recruiting the ULK Complex to Cytosol-Invading Bacteria. Mol Cell 74(2): 320-329 e326. 10.1016/j.molcel.2019.01.041
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N (2009). Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 20(7): 1981-1991. 10.1091/mbc.E08-12-1248
- Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P (2000). Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem 275(2): 992-998. 10.1074/jbc.275.2.992
- Chang C, Young LN, Morris KL, von Bulow S, Schoneberg J, Yamamoto-Imoto H, Oe Y, Yamamoto K, Nakamura S, Stjepanovic G, Hummer G, Yoshimori T, Hurley JH (2019). Bidirectional Control of Autophagy by BECN1 BARA Domain Dynamics. Mol Cell 73(2): 339-353 e336. 10.1016/j.molcel.2018.10.035
- Baskaran S, Carlson LA, Stjepanovic G, Young LN, Kim do J, Grob P, Stanley RE, Nogales E, Hurley JH (2014). Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. Elife 3: e05115. 10.7554/eLife.05115
- He C, Levine B (2010). The Beclin 1 interactome. Curr Opin Cell Biol 22(2): 140-149. 10.1016/j.ceb.2010.01.001
- Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL (2013). Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 152(1-2): 290-303. 10.1016/j.cell.2012.12.016
- Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA (2006). Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 119(Pt 18): 3888-3900. 10.1242/jcs.03172
- Velikkakath AK, Nishimura T, Oita E, Ishihara N, Mizushima N (2012). Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol Biol Cell 23(5): 896-909. 10.1091/mbc.E11-09-0785
- Bakula D, Muller AJ, Zuleger T, Takacs Z, Franz-Wachtel M, Thost AK, Brigger D, Tschan MP, Frickey T, Robenek H, Macek B, Proikas-Cezanne T (2017). WIPI3 and WIPI4 beta-propellers are scaffolds for LKB1-AMPK-TSC signalling circuits in the control of autophagy. Nat Commun 8: 15637. 10.1038/ncomms15637
- Nishimura T, Kaizuka T, Cadwell K, Sahani MH, Saitoh T, Akira S, Virgin HW, Mizushima N (2013). FIP200 regulates targeting of Atg16L1 to the isolation membrane. Embo Rep 14(3): 284-291. 10.1038/embor.2013.6
- Gammoh N, Florey O, Overholtzer M, Jiang X (2013). Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex-dependent and -independent autophagy. Nat Struct Mol Biol 20(2): 144-149. 10.1038/nsmb.2475
- Fujita N, Morita E, Itoh T, Tanaka A, Nakaoka M, Osada Y, Umemoto T, Saitoh T, Nakatogawa H, Kobayashi S, Haraguchi T, Guan JL, Iwai K, Tokunaga F, Saito K, Ishibashi K, Akira S, Fukuda M, Noda T, Yoshimori T (2013). Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J Cell Biol 203(1): 115-128. 10.1083/jcb.201304188
- Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA (2014). WIPI2 Links LC3 Conjugation with PI3P, Autophagosome Formation, and Pathogen Clearance by Recruiting Atg12-5-16L1. Mol Cell 55(2): 238-252. 10.1016/j.molcel.2014.05.021
- Park S, Buck MD, Desai C, Zhang X, Loginicheva E, Martinez J, Freeman ML, Saitoh T, Akira S, Guan JL, He YW, Blackman MA, Handley SA, Levine B, Green DR, Reese TA, Artyomov MN, Virgin HW (2016). Autophagy Genes Enhance Murine Gammaherpesvirus 68 Reactivation from Latency by Preventing Virus-Induced Systemic Inflammation. Cell Host Microbe 19(1): 91-101. 10.1016/j.chom.2015.12.010
- Jao CC, Ragusa MJ, Stanley RE, Hurley JH (2013). A HORMA domain in Atg13 mediates PI 3-kinase recruitment in autophagy. Proc Natl Acad Sci U S A 110(14): 5486-5491. 10.1073/pnas.1220306110
- Itakura E, Kishi-Itakura C, Mizushima N (2012). The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151(6): 1256-1269. 10.1016/j.cell.2012.11.001
- Matsui T, Jiang P, Nakano S, Sakamaki Y, Yamamoto H, Mizushima N (2018). Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. J Cell Biol 217(8):2633-2645. 10.1083/jcb.201712058
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T (2005). p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171(4): 603-614. 10.1083/jcb.200507002
- Turco E, Witt M, Abert C, Bock-Bierbaum T, Su MY, Trapannone R, Sztacho M, Danieli A, Shi X, Zaffagnini G, Gamper A, Schuschnig M, Fracchiolla D, Bernklau D, Romanov J, Hartl M, Hurley JH, Daumke O, Martens S (2019). FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates. Mol Cell 74(2): 330-346 e311. 10.1016/j.molcel.2019.01.035
- Smith MD, Harley ME, Kemp AJ, Wills J, Lee M, Arends M, von Kriegsheim A, Behrends C, Wilkinson S (2018). CCPG1 Is a Non-canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis. Dev Cell 44(2): 217-232 e211. 10.1016/j.devcel.2017.11.024
- Stolz A, Ernst A, Dikic I (2014). Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 16(6): 495-501. 10.1038/ncb2979
- Tsuboyama K, Koyama-Honda I, Sakamaki Y, Koike M, Morishita H, Mizushima N (2016). The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science 354(6315): 1036-1041. 10.1126/science.aaf6136
- Nguyen TN, Padman BS, Usher J, Oorschot V, Ramm G, Lazarou M (2016). Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J Cell Biol 215(6): 857-874. 10.1083/jcb.201607039
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T (2000). LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. Embo J 19(21): 5720-5728. 10.1093/emboj/19.21.5720
- Sagiv Y, Legesse-Miller A, Porat A, Elazar Z (2000). GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. Embo J 19(7): 1494-1504. 10.1093/emboj/19.7.1494
- Kumar S, Jain A, Farzam F, Jia J, Gu Y, Choi SW, Mudd MH, Claude-Taupin A, Wester MJ, Lidke KA, Rusten TE, Deretic V (2018). Mechanism of Stx17 recruitment to autophagosomes via IRGM and mammalian Atg8 proteins. J Cell Biol 217(3): 997-1013. 10.1083/jcb.201708039
- Gu Y, Princely Abudu Y, Kumar S, Bissa B, Choi SW, Jia J, Lazarou M, Eskelinen EL, Johansen T, Deretic V (2019). Mammalian Atg8 proteins regulate lysosome and autolysosome biogenesis through SNAREs. Embo J 38(22): e101994. 10.15252/embj.2019101994
- Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR (2007). Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450(7173): 1253-1257. 10.1038/nature06421
- Heckmann BL, Teubner BJW, Tummers B, Boada-Romero E, Harris L, Yang M, Guy CS, Zakharenko SS, Green DR (2019). LC3-Associated Endocytosis Facilitates beta-Amyloid Clearance and Mitigates Neurodegeneration in Murine Alzheimer's Disease. Cell 178(3): 536-551 e514. 10.1016/j.cell.2019.05.056
- Florey O, Gammoh N, Kim SE, Jiang X, Overholtzer M (2015). V-ATPase and osmotic imbalances activate endolysosomal LC3 lipidation. Autophagy 11(1): 88-99. 10.4161/15548627.2014.984277
- Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M (2011). Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol 13(11): 1335-1343. 10.1038/ncb2363
- Jacquin E, Leclerc-Mercier S, Judon C, Blanchard E, Fraitag S, Florey O (2017). Pharmacological modulators of autophagy activate a parallel noncanonical pathway driving unconventional LC3 lipidation. Autophagy 13(5): 854-867. 10.1080/15548627.2017.1287653
- Fletcher K, Ulferts R, Jacquin E, Veith T, Gammoh N, Arasteh JM, Mayer U, Carding SR, Wileman T, Beale R, Florey O (2018). The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. Embo J 37(4). 10.15252/embj.201797840
- Rai S, Arasteh M, Jefferson M, Pearson T, Wang Y, Zhang W, Bicsak B, Divekar D, Powell PP, Naumann R, Beraza N, Carding SR, Florey O, Mayer U, Wileman T (2019). The ATG5-binding and coiled coil domains of ATG16L1 maintain autophagy and tissue homeostasis in mice independently of the WD domain required for LC3-associated phagocytosis. Autophagy 15(4): 599-612. 10.1080/15548627.2018.1534507
- Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, Hengartner MO, Green DR (2011). Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci U S A 108(42): 17396-17401. 10.1073/pnas.1113421108
- Heckmann BL, Green DR (2019). LC3-associated phagocytosis at a glance. J Cell Sci 132(5): jcs222984. 10.1242/jcs.222984
- Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V (2008). Toll-like receptors control autophagy. Embo J 27(7): 1110-1121. 10.1038/emboj.2008.31
- Martinez J, Malireddi RK, Lu Q, Cunha LD, Pelletier S, Gingras S, Orchard R, Guan JL, Tan H, Peng J, Kanneganti TD, Virgin HW, Green DR (2015). Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol 17(7): 893-906. 10.1038/ncb3192
- Lambeth JD (2004). NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol 4(3): 181-189. 10.1038/nri1312
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z (2007). Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. Embo J 26(7): 1749-1760. 10.1038/sj.emboj.7601623
- Martinez J, Cunha LD, Park S, Yang M, Lu Q, Orchard R, Li QZ, Yan M, Janke L, Guy C, Linkermann A, Virgin HW, Green DR (2016). Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature 533(7601): 115-119. 10.1038/nature17950
- Kageyama S, Omori H, Saitoh T, Sone T, Guan JL, Akira S, Imamoto F, Noda T, Yoshimori T (2011). The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol Biol Cell 22(13): 2290-2300. 10.1091/mbc.E10-11-0893
- Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, Hengartner MO, Green DR (2011). Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci U S A 108(42): 17396-17401. 10.1073/pnas.1113421108
- Boada-Romero E, Martinez J, Heckmann BL, Green DR (2020). The clearance of dead cells by efferocytosis. Nat Rev Mol Cell Biol 21(7): 398-414. 10.1038/s41580-020-0232-1
- Kim JY, Zhao H, Martinez J, Doggett TA, Kolesnikov AV, Tang PH, Ablonczy Z, Chan CC, Zhou Z, Green DR, Ferguson TA (2013). Noncanonical autophagy promotes the visual cycle. Cell 154(2): 365-376. 10.1016/j.cell.2013.06.012
- Cunha LD, Yang M, Carter R, Guy C, Harris L, Crawford JC, Quarato G, Boada-Romero E, Kalkavan H, Johnson MDL, Natarajan S, Turnis ME, Finkelstein D, Opferman JT, Gawad C, Green DR (2018). LC3-Associated Phagocytosis in Myeloid Cells Promotes Tumor Immune Tolerance. Cell 175(2): 429-441 e416. 10.1016/j.cell.2018.08.061
- Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, Sasai M, Latz E, Brinkmann MM, Iwasaki A, Coyle AJ, Kolbeck R, Green DR, Sanjuan MA (2012). Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity 37(6): 986-997. 10.1016/j.immuni.2012.09.014
- Lee C, Lamech L, Johns E, Overholtzer M (2020). Selective Lysosome Membrane Turnover Is Induced by Nutrient Starvation. Dev Cell 55(3): 289-297 e284. 10.1016/j.devcel.2020.08.008
- De Duve C, Wattiaux R (1966). Functions of lysosomes. Annu Rev Physiol 28: 435-492. 10.1146/annurev.ph.28.030166.002251
- Li WW, Li J, Bao JK (2012). Microautophagy: lesser-known self-eating. Cell Mol Life Sci 69(7): 1125-1136. 10.1007/s00018-011-0865-5
- Oku M, Sakai Y (2018). Three Distinct Types of Microautophagy Based on Membrane Dynamics and Molecular Machineries. Bioessays 40(6): e1800008. 10.1002/bies.201800008
- Schuck S (2020). Microautophagy – distinct molecular mechanisms handle cargoes of many sizes. J Cell Sci 133(17): jcs246322. 10.1242/jcs.246322
- Loi M, Raimondi A, Morone D, Molinari M (2019). ESCRT-III-driven piecemeal micro-ER-phagy remodels the ER during recovery from ER stress. Nat Commun 10(1): 5058. 10.1038/s41467-019-12991-z
- Mejlvang J, Olsvik H, Svenning S, Bruun JA, Abudu YP, Larsen KB, Brech A, Hansen TE, Brenne H, Hansen T, Stenmark H, Johansen T (2018). Starvation induces rapid degradation of selective autophagy receptors by endosomal microautophagy. J Cell Biol 217(10):3640-3655. 10.1083/jcb.201711002
- Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N (2008). FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 181(3): 497-510. 10.1083/jcb.200712064
- Raposo G, Stoorvogel W (2013). Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200(4): 373-383. 10.1083/jcb.201211138
- Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, et al. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 7(1): 1535750. 10.1080/20013078.2018.1535750
- van Niel G, D'Angelo G, Raposo G (2018). Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 19(4): 213-228. 10.1038/nrm.2017.125
- Raposo G, Stahl PD (2019). Extracellular vesicles: a new communication paradigm? Nat Rev Mol Cell Biol 20(9): 509-510. 10.1038/s41580-019-0158-7
- van der Pol E, Boing AN, Harrison P, Sturk A, Nieuwland R (2012). Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev 64(3): 676-705. 10.1124/pr.112.005983
- Pitt JM, Kroemer G, Zitvogel L (2016). Extracellular vesicles: masters of intercellular communication and potential clinical interventions. J Clin Invest 126(4): 1139-1143. 10.1172/JCI87316
- Maas SLN, Breakefield XO, Weaver AM (2017). Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol 27(3): 172-188. 10.1016/j.tcb.2016.11.003
- Murrow L, Malhotra R, Debnath J (2015). ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat Cell Biol 17(3): 300-310. 10.1038/ncb3112
- Guo H, Chitiprolu M, Roncevic L, Javalet C, Hemming FJ, Trung MT, Meng L, Latreille E, Tanese de Souza C, McCulloch D, Baldwin RM, Auer R, Cote J, Russell RC, Sadoul R, Gibbings D (2017). Atg5 Disassociates the V1V0-ATPase to Promote Exosome Production and Tumor Metastasis Independent of Canonical Macroautophagy. Dev Cell 43(6): 716-730 e717. 10.1016/j.devcel.2017.11.018
- Leidal AM, Huang HH, Marsh T, Solvik T, Zhang D, Ye J, Kai F, Goldsmith J, Liu JY, Huang YH, Monkkonen T, Vlahakis A, Huang EJ, Goodarzi H, Yu L, Wiita AP, Debnath J (2020). The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat Cell Biol 22(2): 187-199. 10.1038/s41556-019-0450-y
- Keller MD, Ching KL, Liang FX, Dhabaria A, Tam K, Ueberheide BM, Unutmaz D, Torres VJ, Cadwell K (2020). Decoy exosomes provide protection against bacterial toxins. Nature 579(7798): 260-264. 10.1038/s41586-020-2066-6
- Ponpuak M, Mandell MA, Kimura T, Chauhan S, Cleyrat C, Deretic V (2015). Secretory autophagy. Curr Opin Cell Biol 35: 106-116. 10.1016/j.ceb.2015.04.016
- Claude-Taupin A, Jia J, Mudd M, Deretic V (2017). Autophagy's secret life: secretion instead of degradation. Essays Biochem 61(6): 637-647. 10.1042/EBC20170024
- Gerstenmaier L, Pilla R, Herrmann L, Herrmann H, Prado M, Villafano GJ, Kolonko M, Reimer R, Soldati T, King JS, Hagedorn M (2015). The autophagic machinery ensures nonlytic transmission of mycobacteria. Proc Natl Acad Sci U S A 112(7):E687-92. 10.1073/pnas.1423318112
- Robinson SM, Tsueng G, Sin J, Mangale V, Rahawi S, McIntyre LL, Williams W, Kha N, Cruz C, Hancock BM, Nguyen DP, Sayen MR, Hilton BJ, Doran KS, Segall AM, Wolkowicz R, Cornell CT, Whitton JL, Gottlieb RA, Feuer R (2014). Coxsackievirus B exits the host cell in shed microvesicles displaying autophagosomal markers. PLoS pathogens 10(4): e1004045. 10.1371/journal.ppat.1004045
- Beale R, Wise H, Stuart A, Ravenhill BJ, Digard P, Randow F (2014). A LC3-interacting motif in the influenza A virus M2 protein is required to subvert autophagy and maintain virion stability. Cell Host Microbe 15(2): 239-247. 10.1016/j.chom.2014.01.006
- Kimura T, Jia J, Kumar S, Choi SW, Gu Y, Mudd M, Dupont N, Jiang S, Peters R, Farzam F, Jain A, Lidke KA, Adams CM, Johansen T, Deretic V (2017). Dedicated SNAREs and specialized TRIM cargo receptors mediate secretory autophagy. Embo J 36(1): 42-60. 10.15252/embj.201695081
- Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V (2011). Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. Embo J 30(23): 4701-4711. 10.1038/emboj.2011.398
- Razani B, Feng C, Coleman T, Emanuel R, Wen H, Hwang S, Ting JP, Virgin HW, Kastan MB, Semenkovich CF (2012). Autophagy links inflammasomes to atherosclerotic progression. Cell Metab 15(4): 534-544. 10.1016/j.cmet.2012.02.011
- Karmakar M, Minns M, Greenberg EN, Diaz-Aponte J, Pestonjamasp K, Johnson JL, Rathkey JK, Abbott DW, Wang K, Shao F, Catz SD, Dubyak GR, Pearlman E (2020). N-GSDMD trafficking to neutrophil organelles facilitates IL-1beta release independently of plasma membrane pores and pyroptosis. Nat Commun 11(1): 2212. 10.1038/s41467-020-16043-9
- Rubartelli A, Cozzolino F, Talio M, Sitia R (1990). A novel secretory pathway for interleukin-1 beta, a protein lacking a signal sequence. Embo J 9(5): 1503-1510. 10.1002/j.1460-2075.1990.tb08268.x
- Lock R, Kenific CM, Leidal AM, Salas E, Debnath J (2014). Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer Discov 4(4): 466-479. 10.1158/2159-8290.CD-13-0841
- Thorburn J, Horita H, Redzic J, Hansen K, Frankel AE, Thorburn A (2009). Autophagy regulates selective HMGB1 release in tumor cells that are destined to die. Cell Death Diff 16(1): 175-183. 10.1038/cdd.2008.143
- Zhang M, Kenny S, Ge L, Xu K, Schekman R (2015). Translocation of interleukin-1beta into a vesicle intermediate in autophagy-mediated secretion. Elife 4: e11205. 10.7554/eLife.11205
- Zhang M, Liu L, Lin X, Wang Y, Li Y, Guo Q, Li S, Sun Y, Tao X, Zhang D, Lv X, Zheng L, Ge L (2020). A Translocation Pathway for Vesicle-Mediated Unconventional Protein Secretion. Cell 181(3): 637-652 e615. 10.1016/j.cell.2020.03.031
- Broz P, Pelegrin P, Shao F (2020). The gasdermins, a protein family executing cell death and inflammation. Nat Rev Immunol 20(3): 143-157. 10.1038/s41577-019-0228-2
- Evavold CL, Ruan J, Tan Y, Xia S, Wu H, Kagan JC (2018). The Pore-Forming Protein Gasdermin D Regulates Interleukin-1 Secretion from Living Macrophages. Immunity 48(1): 35-44 e36. 10.1016/j.immuni.2017.11.013
- Zhang M, Schekman R (2013). Cell biology. Unconventional secretion, unconventional solutions. Science 340(6132): 559-561. 10.1126/science.1234740
- Holthuis JC, Menon AK (2014). Lipid landscapes and pipelines in membrane homeostasis. Nature 510(7503): 48-57. 10.1038/nature13474
- Simons K (2016). Cell membranes: A subjective perspective. Biochim Biophys Acta 1858(10): 2569-2572. 10.1016/j.bbamem.2016.01.023
- Tang SKY, Marshall WF (2017). Self-repairing cells: How single cells heal membrane ruptures and restore lost structures. Science 356(6342): 1022-1025. 10.1126/science.aam6496
- Izzo V, Bravo-San Pedro JM, Sica V, Kroemer G, Galluzzi L (2016). Mitochondrial Permeability Transition: New Findings and Persisting Uncertainties. Trends Cell Biol 26(9): 655-667. 10.1016/j.tcb.2016.04.006
- Werneburg NW, Guicciardi ME, Bronk SF, Kaufmann SH, Gores GJ (2007). Tumor necrosis factor-related apoptosis-inducing ligand activates a lysosomal pathway of apoptosis that is regulated by Bcl-2 proteins. J Biol Chem 282(39): 28960-28970. 10.1074/jbc.M705671200
- Rapoport SI, Hori M, Klatzo I (1971). Reversible osmotic opening of the blood-brain barrier. Science 173(4001): 1026-1028. 10.1126/science.173.4001.1026
- Tan JMJ, Mellouk N, Osborne SE, Ammendolia DA, Dyer DN, Li R, Brunen D, van Rijn JM, Huang J, Czuczman MA, Cemma MA, Won AM, Yip CM, Xavier RJ, MacDuff DA, Reggiori F, Debnath J, Yoshimori T, Kim PK, Fairn GD, Coyaud E, Raught B, Muise AM, Higgins DE, Brumell JH (2018). An ATG16L1-dependent pathway promotes plasma membrane repair and limits Listeria monocytogenes cell-to-cell spread. Nat Microbiol 3(12): 1472-1485. 10.1038/s41564-018-0293-5
- Sonder SL, Hager SC, Heitmann ASB, Frankel LB, Dias C, Simonsen AC, Nylandsted J (2021). Restructuring of the plasma membrane upon damage by LC3-associated macropinocytosis. Sci Adv 7(27): eabg1969. 10.1126/sciadv.abg1969
- Claude-Taupin A, Jia J, Bhujabal Z, Garfa-Traore M, Kumar S, da Silva GPD, Javed R, Gu Y, Allers L, Peters R, Wang F, da Costa LJ, Pallikkuth S, Lidke KA, Mauthe M, Verlhac P, Uchiyama Y, Salemi M, Phinney B, Tooze SA, Mari MC, Johansen T, Reggiori F, Deretic V (2021). ATG9A protects the plasma membrane from programmed and incidental permeabilization. Nat Cell Biol. 10.1038/s41556-021-00706-w
- Ballabio A, Bonifacino JS (2020). Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat Rev Mol Cell Biol 21(2): 101-118. 10.1038/s41580-019-0185-4
- Saftig P, Puertollano R (2021). How Lysosomes Sense, Integrate, and Cope with Stress. Trends Biochem Sci 46(2): 97-112. 10.1016/j.tibs.2020.09.004
- Lu S, Sung T, Lin N, Abraham RT, Jessen BA (2017). Lysosomal adaptation: How cells respond to lysosomotropic compounds. PLoS One 12(3): e0173771. 10.1371/journal.pone.0173771
- Rusmini P, Cortese K, Crippa V, Cristofani R, Cicardi ME, Ferrari V, Vezzoli G, Tedesco B, Meroni M, Messi E, Piccolella M, Galbiati M, Garre M, Morelli E, Vaccari T, Poletti A (2019). Trehalose induces autophagy via lysosomal-mediated TFEB activation in models of motoneuron degeneration. Autophagy 15(4): 631-651. 10.1080/15548627.2018.1535292
- Flavin WP, Bousset L, Green ZC, Chu Y, Skarpathiotis S, Chaney MJ, Kordower JH, Melki R, Campbell EM (2017). Endocytic vesicle rupture is a conserved mechanism of cellular invasion by amyloid proteins. Acta Neuropathol 134(4): 629-653. 10.1007/s00401-017-1722-x
- Papadopoulos C, Kirchner P, Bug M, Grum D, Koerver L, Schulze N, Poehler R, Dressler A, Fengler S, Arhzaouy K, Lux V, Ehrmann M, Weihl CC, Meyer H (2017). VCP/p97 cooperates with YOD1, UBXD1 and PLAA to drive clearance of ruptured lysosomes by autophagy. Embo J 36(2): 135-150. 10.15252/embj.201695148
- Jia J, Claude-Taupin A, Gu Y, Choi SW, Peters R, Bissa B, Mudd MH, Allers L, Pallikkuth S, Lidke KA, Salemi M, Phinney B, Mari M, Reggiori F, Deretic V (2020). Galectin-3 Coordinates a Cellular System for Lysosomal Repair and Removal. Dev Cell 52(1): 69-87 e68. 10.1016/j.devcel.2019.10.025
- Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E (2008). Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol 9(8): 847-856. 10.1038/ni.1631
- Maejima I, Takahashi A, Omori H, Kimura T, Takabatake Y, Saitoh T, Yamamoto A, Hamasaki M, Noda T, Isaka Y, Yoshimori T (2013). Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. Embo J 32(17): 2336-2347. 10.1038/emboj.2013.171
- Chen X, Khambu B, Zhang H, Gao W, Li M, Chen X, Yoshimori T, Yin XM (2014). Autophagy induced by calcium phosphate precipitates targets damaged endosomes. J Biol Chem 289(16): 11162-11174. 10.1074/jbc.M113.531855
- Perrin AJ, Jiang X, Birmingham CL, So NS, Brumell JH (2004). Recognition of bacteria in the cytosol of Mammalian cells by the ubiquitin system. Curr Biol 14(9): 806-811. 10.1074/jbc.M113.531855
- Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F (2012). Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482(7385): 414-418. 10.1038/nature10744
- Herrero-Martin G, Hoyer-Hansen M, Garcia-Garcia C, Fumarola C, Farkas T, Lopez-Rivas A, Jaattela M (2009). TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. Embo J 28(6): 677-685. 10.1038/emboj.2009.8
- Okada M, Matsuzawa A, Yoshimura A, Ichijo H (2014). The lysosome rupture-activated TAK1-JNK pathway regulates NLRP3 inflammasome activation. J Biol Chem 289(47): 32926-32936. 10.1074/jbc.M114.579961
- Sakamachi Y, Morioka S, Mihaly SR, Takaesu G, Foley JF, Fessler MB, Ninomiya-Tsuji J (2017). TAK1 regulates resident macrophages by protecting lysosomal integrity. Cell Death Dis 8(2): e2598. 10.1038/cddis.2017.23
- Jia J, Bissa B, Brecht L, Allers L, Choi SW, Gu Y, Zbinden M, Burge MR, Timmins G, Hallows K, Behrends C, Deretic V (2020). AMPK, a Regulator of Metabolism and Autophagy, Is Activated by Lysosomal Damage via a Novel Galectin-Directed Ubiquitin Signal Transduction System. Mol Cell. 10.1016/j.molcel.2019.12.028
- Chauhan S, Kumar S, Jain A, Ponpuak M, Mudd MH, Kimura T, Choi SW, Peters R, Mandell M, Bruun JA, Johansen T, Deretic V (2016). TRIMs and Galectins Globally Cooperate and TRIM16 and Galectin-3 Co-direct Autophagy in Endomembrane Damage Homeostasis. Dev Cell 39(1): 13-27. 10.1016/j.devcel.2016.08.003
- Koerver L, Papadopoulos C, Liu B, Kravic B, Rota G, Brecht L, Veenendaal T, Polajnar M, Bluemke A, Ehrmann M, Klumperman J, Jaattela M, Behrends C, Meyer H (2019). The ubiquitin-conjugating enzyme UBE2QL1 coordinates lysophagy in response to endolysosomal damage. Embo Rep 20(10): e48014. 10.15252/embr.201948014
- Yoshida Y, Yasuda S, Fujita T, Hamasaki M, Murakami A, Kawawaki J, Iwai K, Saeki Y, Yoshimori T, Matsuda N, Tanaka K (2017). Ubiquitination of exposed glycoproteins by SCF(FBXO27) directs damaged lysosomes for autophagy. Proc Natl Acad Sci U S A 114(32): 8574-8579. 10.1073/pnas.1702615114
- Skowyra ML, Schlesinger PH, Naismith TV, Hanson PI (2018). Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 360(6384): eaar5078. 10.1126/science.aar5078
- Radulovic M, Schink KO, Wenzel EM, Nahse V, Bongiovanni A, Lafont F, Stenmark H (2018). ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival. Embo J 37(21): e99753. 10.15252/embj.201899753
- Gupta S, Yano J, Mercier V, Htwe HH, Shin HR, Rademaker G, Cakir Z, Ituarte T, Wen KW, Kim GE, Zoncu R, Roux A, Dawson DW, Perera RM (2021). Lysosomal retargeting of Myoferlin mitigates membrane stress to enable pancreatic cancer growth. Nat Cell Biol 23(3): 232-242. 10.1038/s41556-021-00644-7
- Weber RA, Yen FS, Nicholson SPV, Alwaseem H, Bayraktar EC, Alam M, Timson RC, La K, Abu-Remaileh M, Molina H, Birsoy K (2020). Maintaining Iron Homeostasis Is the Key Role of Lysosomal Acidity for Cell Proliferation. Mol Cell 77(3): 645-655 e647. 10.1016/j.molcel.2020.01.003
- Jia J, Abudu YP, Claude-Taupin A, Gu Y, Kumar S, Choi SW, Peters R, Mudd MH, Allers L, Salemi M, Phinney B, Johansen T, Deretic V (2018). Galectins Control mTOR in Response to Endomembrane Damage. Mol Cell 70(1): 120-135 e128. 10.1016/j.molcel.2018.03.009
- Paz I, Sachse M, Dupont N, Mounier J, Cederfur C, Enninga J, Leffler H, Poirier F, Prevost MC, Lafont F, Sansonetti P (2010). Galectin-3, a marker for vacuole lysis by invasive pathogens. Cell Microbiol 12(4): 530-544. 10.1111/j.1462-5822.2009.01415.x
- Aits S, Kricker J, Liu B, Ellegaard AM, Hamalisto S, Tvingsholm S, Corcelle-Termeau E, Hogh S, Farkas T, Holm Jonassen A, Gromova I, Mortensen M, Jaattela M (2015). Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy 11(8): 1408-1424. 10.1080/15548627.2015.1063871
- Eapen V, Swarup S, Hoyer M, Paolo J, Harper J Quantitative proteomics reveals the selectivity of ubiquitin-binding autophagy receptors in the turnover of damaged lysosomes by lysophagy. BioRxiv. 10.1101/2021.07.19.452535
- Kumar S, Jain A, Choi SW, da Silva GPD, Allers L, Mudd MH, Peters RS, Anonsen JH, Rusten TE, Lazarou M, Deretic V (2020). Mammalian Atg8 proteins and the autophagy factor IRGM control mTOR and TFEB at a regulatory node critical for responses to pathogens. Nat Cell Biol 22(8): 973-985. 10.1038/s41556-020-0549-1
- Nakamura S, Shigeyama S, Minami S, Shima T, Akayama S, Matsuda T, Esposito A, Napolitano G, Kuma A, Namba-Hamano T, Nakamura J, Yamamoto K, Sasai M, Tokumura A, Miyamoto M, Oe Y, Fujita T, Terawaki S, Takahashi A, Hamasaki M, Yamamoto M, Okada Y, Komatsu M, Nagai T, Takabatake Y, Xu H, Isaka Y, Ballabio A, Yoshimori T (2020). LC3 lipidation is essential for TFEB activation during the lysosomal damage response to kidney injury. Nat Cell Biol 22(10): 1252-1263. 10.1038/s41556-020-00583-9
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A (2009). A gene network regulating lysosomal biogenesis and function. Science 325(5939): 473-477. 10.1126/science.1174447
- Palmieri M, Impey S, Kang HJ, di Ronza A, Pelz C, Sardiello M, Ballabio A (2011). Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet 20(19): 3852-3866. 10.1093/hmg/ddr306
- Napolitano G, Ballabio A (2016). TFEB at a glance. J Cell Sci 129(13): 2475-2481. 10.1242/jcs.146365
- Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, Wollenberg AC, Di Bernardo D, Chan L, Irazoqui JE, Ballabio A (2013). TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol 15(6): 647-658. 10.1038/ncb2718
- Medina DL, Fraldi A, Bouche V, Annunziata F, Mansueto G, Spampanato C, Puri C, Pignata A, Martina JA, Sardiello M, Palmieri M, Polishchuk R, Puertollano R, Ballabio A (2011). Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell 21(3): 421-430. 10.1016/j.devcel.2011.07.016
- Mansueto G, Armani A, Viscomi C, D'Orsi L, De Cegli R, Polishchuk EV, Lamperti C, Di Meo I, Romanello V, Marchet S, Saha PK, Zong H, Blaauw B, Solagna F, Tezze C, Grumati P, Bonaldo P, Pessin JE, Zeviani M, Sandri M, Ballabio A (2017). Transcription Factor EB Controls Metabolic Flexibility during Exercise. Cell Metab 25(1): 182-196. 10.1016/j.cmet.2016.11.003
- Nezich CL, Wang C, Fogel AI, Youle RJ (2015). MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J Cell Biol 210(3): 435-450. 10.1083/jcb.201501002
- Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM (2012). The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 5(228): ra42. 10.1126/scisignal.2002790
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A (2012). A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. Embo J 31(5): 1095-1108. 10.1038/emboj.2012.32
- Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, Settembre C, Wang W, Gao Q, Xu H, Sandri M, Rizzuto R, De Matteis MA, Ballabio A (2015). Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 17(3): 288-299. 10.1038/ncb3114
- Napolitano G, Di Malta C, Esposito A, de Araujo MEG, Pece S, Bertalot G, Matarese M, Benedetti V, Zampelli A, Stasyk T, Siciliano D, Venuta A, Cesana M, Vilardo C, Nusco E, Monfregola J, Calcagni A, Di Fiore PP, Huber LA, Ballabio A (2020). A substrate-specific mTORC1 pathway underlies Birt-Hogg-Dube syndrome. Nature 585(7826): 597-602. 10.1038/s41586-020-2444-0
- Visvikis O, Ihuegbu N, Labed SA, Luhachack LG, Alves AF, Wollenberg AC, Stuart LM, Stormo GD, Irazoqui JE (2014). Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity 40(6): 896-909. 10.1016/j.immuni.2014.05.002
- Campbell GR, Rawat P, Bruckman RS, Spector SA (2015). Human Immunodeficiency Virus Type 1 Nef Inhibits Autophagy through Transcription Factor EB Sequestration. PLoS Pathog 11(6): e1005018. 10.1371/journal.ppat.1005018
- Najibi M, Labed SA, Visvikis O, Irazoqui JE (2016). An Evolutionarily Conserved PLC-PKD-TFEB Pathway for Host Defense. Cell Rep 15(8): 1728-1742. 10.1016/j.celrep.2016.04.052
- El-Houjeiri L, Possik E, Vijayaraghavan T, Paquette M, Martina JA, Kazan JM, Ma EH, Jones R, Blanchette P, Puertollano R, Pause A (2019). The Transcription Factors TFEB and TFE3 Link the FLCN-AMPK Signaling Axis to Innate Immune Response and Pathogen Resistance. Cell Rep 26(13): 3613-3628 e3616. 10.1016/j.celrep.2019.02.102
- Goodwin JMF, O; Murphy, L.O (2020). GABARAP membrane conjugation sequesters the FLCN-FNIP tumor suppressor complex to activate TFEB and lysosomal biogenesis. BioxRiv. 10.1101/2021.02.22.432209
- Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM (2013). The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell 52(4): 495-505. 10.1016/j.molcel.2013.09.016
- Meng J, Ferguson SM (2018). GATOR1-dependent recruitment of FLCN-FNIP to lysosomes coordinates Rag GTPase heterodimer nucleotide status in response to amino acids. J Cell Biol 217(8): 2765-2776. 10.1083/jcb.201712177
- Otten EG, Werner E, Crespillo-Casado A, Boyle KB, Dharamdasani V, Pathe C, Santhanam B, Randow F (2021). Ubiquitylation of lipopolysaccharide by RNF213 during bacterial infection. Nature 594(7861):111-116. 10.1038/s41586-021-03566-4
- Agrotis A, von Chamier L, Oliver H, Kiso K, Singh T, Ketteler R (2019). Human ATG4 autophagy proteases counteract attachment of ubiquitin-like LC3/GABARAP proteins to other cellular proteins. J Biol Chem 294(34): 12610-12621. 10.1074/jbc.AC119.009977
- Nguyen TN, Padman BS, Zellner S, Khuu G, Uoselis L, Lam WK, Skulsuppaisarn M, Lindblom RSJ, Watts EM, Behrends C, Lazarou M (2021). ATG4 family proteins drive phagophore growth independently of the LC3/GABARAP lipidation system. Mol Cell 81(9): 2013-2030 e2019. 10.1016/j.molcel.2021.03.001
- Levine B, Kroemer G (2019). Biological Functions of Autophagy Genes: A Disease Perspective. Cell 176(1-2): 11-42. 10.1016/j.cell.2018.09.048
- Deretic V (2021). Autophagy in inflammation, infection, and immunometabolism. Immunity 54(3): 437-453. 10.1016/j.immuni.2021.01.018
–
ACKNOWLEDGMENTS
We thank the reviewers for their comments, which are reflected in the open questions at the end of the coda section. This work was supported by NIH grants R37AI042999 and R01AI111935 and center grant P20GM121176 to V.D., and AIM center pilot pro-jects to S.K. and J.J.
COPYRIGHT
© 2021

Atg8ylation as a general membrane stress and remodeling response by Kumar et al. is licensed under a Creative Commons Attribution 4.0 International License.