Back to article: The role of hydrophobic matching on transmembrane helix packing in cells
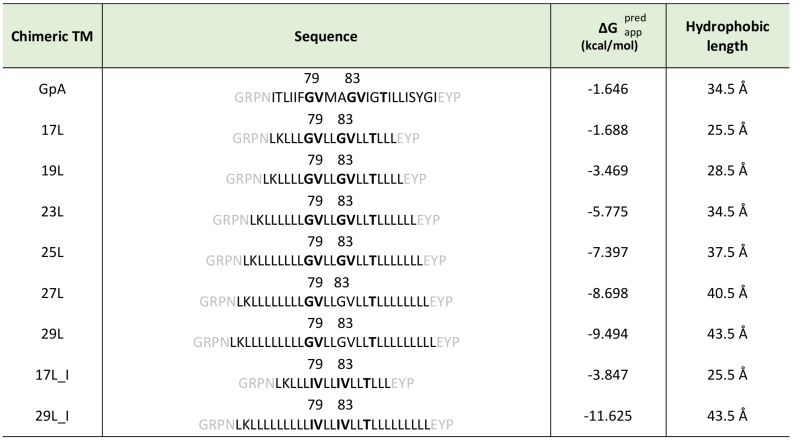
Table 1. Sequences predicted ΔG and hydrophobic length of transmembrane segments.
Chimeric TM segments were named based on their hydrophobic length (amino acids). The sequence of each TM segment is included. The dimerization motive is highlighted in bold (including the amino acid position in the wild-type GpA sequence) and flanking regions are indicated in gray. The apparent predicted ΔG for the insertion of the hydrophobic regions (calculated by the ΔG prediction server v1.0, where negative values are indicative of insertion) and the hydrophobic length (calculated assuming 1.5 Å per residue in a α-helix conformation) were also included in the table.