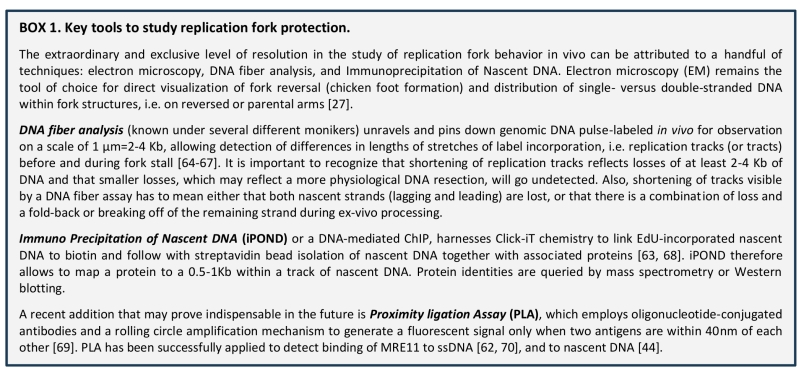
Back to article: A game of substrates: replication fork remodeling and its roles in genome stability and chemo-resistance
27. Vindigni A, Lopes M (2017). Combining electron microscopy with single molecule DNA fiber approaches to study DNA replication dynamics. Biophys Chem 225:3-9. http://dx.doi.org/10.1016/j.bpc.2016.11.014
44. Taglialatela A, Alvarez S, Leuzzi G, Sannino V, Ranjha L, Huang JW, Madubata C, Anand R, Levy B, Rabadan R, Cejka P, Costanzo V, Ciccia A (2017). Restoration of Replication Fork Stability in BRCA1- and BRCA2-Deficient Cells by Inactivation of SNF2-Family Fork Remodelers. Mol Cell 68(2): 414-430. http://dx.doi.org/10.1016/j.molcel.2017.09.036
62. Iannascoli C, Palermo V, Murfuni I, Franchitto A, Pichierri P (2015). The WRN exonuclease domain protects nascent strands from pathological MRE11/EXO1-dependent degradation. Nucleic Acids Res 43(20): 9788-9803. http://dx.doi.org/10.1093/nar/gkv836
63. Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D (2011). Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev 25(12): 1320-1327. http://dx.doi.org/10.1101/gad.2053211
64. Jackson DA, Pombo A (1998). Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 140(6): 1285-1295 https://www.ncbi.nlm.nih.gov/pubmed/?term=9508763
65. Herrick J, Bensimon A (1999). Single molecule analysis of DNA replication. Biochimie 81(8-9): 859-871.https://www.ncbi.nlm.nih.gov/pubmed/?term=10572299
66. Norio P, Schildkraut CL (2001). Visualization of DNA Replication on Individual Epstein-Barr Virus Epi-somes. Science 294(5550): 2361-2364. http://dx.doi.org/10.1126/science.1064603
67. Sidorova JM, Li N, Schwartz DC, Folch A, Monnat RJ, Jr (2009). Microfluidic-assisted analysis of replicating DNA molecules. Nat Protoc 4(6): 849-861. http://dx.doi.org/10.1038/nprot.2009.54
68. Kliszczak AE, Rainey MD, Harhen B, Boisvert FM, Santocanale C (2011). DNA mediated chromatin pull-down for the study of chromatin replication. Sci Rep 1(95): 95. http://dx.doi.org/10.1038/srep00095
69. Weibrecht I, Leuchowius K-J, Clausson C-M, Conze T, Jarvius M, Howell WM, Kamali-Moghaddam M, Söderberg O (2010). Proximity ligation assays: a recent addition to the proteomics toolbox. Expert Rev Proteomics 7(3): 401-409. http://dx.doi.org/10.1586/epr.10.10
70. Palermo V, Rinalducci S, Sanchez M, Grillini F, Sommers JA, Brosh Jr RM, Zolla L, Franchitto A, Pichierri P (2016). CDK1 phosphorylates WRN at collapsed replication forks. Nat Commun 7: 12880. http://dx.doi.org/10.1038/ncomms12880