Back to article: miR34a: a master regulator in the pathogenesis of bronchopulmonary dysplasia
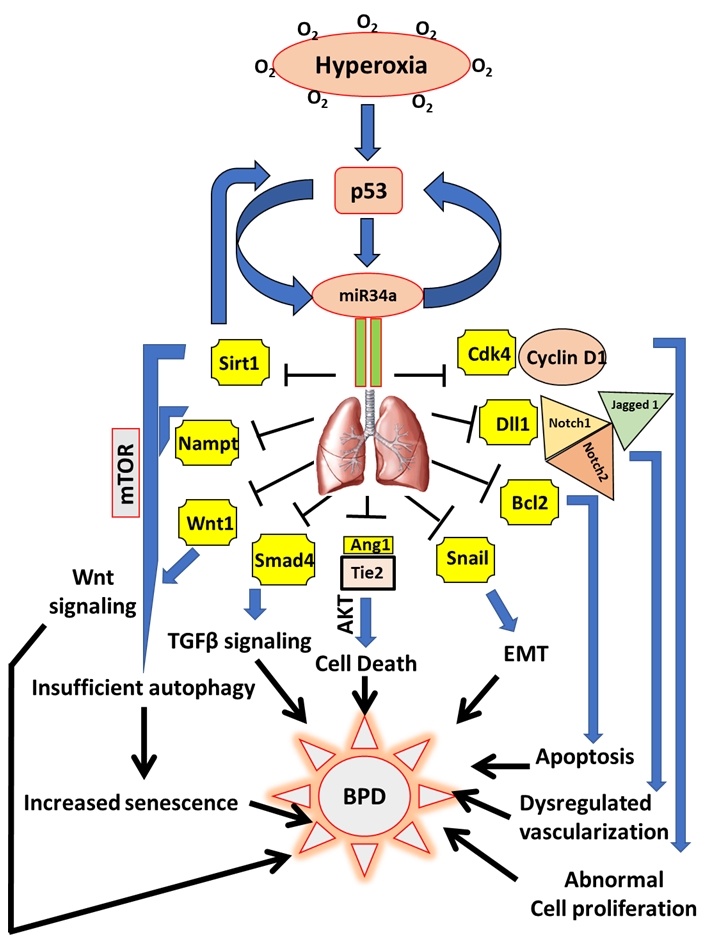
FIGURE 1: miR34a as a master regulator of the proposed cell signaling pathways involved in the pathogenesis of BPD. Hyperoxia exposure to the developing lungs stimulates p53 to activate miR34a in a positive feedback loop. In the lungs, miR34a targets a host of genes which exert their effects either singly or in combination with other genes and signaling pathways that eventually causes BPD in neonates. Upon being targeted by miR34a, Sirt1 is decreased, which leads to an increase in mTOR (a negative regulator of autophagy), that results in insufficient autophagy causing enhanced senescence and impaired alveolarization. Decrease in Sirt1 also increases p53 in a negative feedback loop. Nampt is another target of miR34a which may act in association with Sirt1 to induce senescence. Decrease in Wnt1 and Smad4 interferes with the Wnt and TGFβ signaling pathways responsible for overall lung development/response to hyperoxia. By downregulating Ang1 and its receptor, Tie2, miR34 induces cell death through the Akt pathway. Epithelial-mesenchymal transition (EMT) is brought about by the inhibition of Snail 1, apoptosis by Bcl2, impaired vascularization through inhibition of Dll1 and its receptors Notch1, Notch2 and Jagged 1, and abnormal cell proliferation by inhibition of the Cyclin1-Cdk4 complex. All the above pathophysiological processes that are targeted by miR34a are involved in the pathogenesis of BPD.
Ang1: angiopoetin1; BPD: bronchopulmonary dysplasia; miR: microRNA; mTOR: mechanistic target of Rapamy-cin; Nampt: Nicotinamide phosphoribosyltransferase; Sirt1: Sirtuin1; TGFβ: transforming growth factor beta.