Back to article: The emerging role of minor intron splicing in neurological disorders
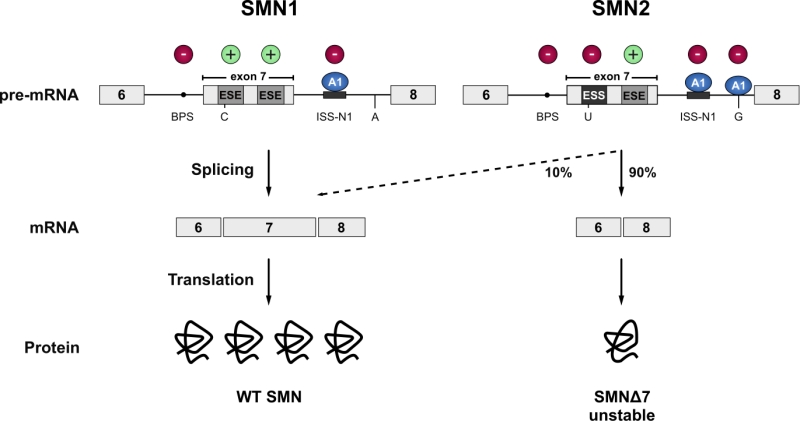
FIGURE 2: SMN1 and SMN2. The vast majority of the SMN protein is produced from the SMN1 gene. However, during evolution, humans have acquired a paralogue (SMN2) by gene duplication, but SMN2 produces only approximately 10% of the full-length mRNA. Due to a C to T transition in SMN2 (C to U at RNA level) the first exonic splicing enhancer (ESE) in exon 7 is disrupted and an exonic splicing silencer (ESS) is created, which is bound by hnRNPA1 (A1). Inclusion of exon 7 is further prevented by an A to G transition further downstream in intron 7 (creating an additional hnRNPA1 binding site), and the suboptimal branchpoint (BPS) in intron 6. Hence, exon 7 is mainly skipped leading to the production of an instable C-terminally truncated protein (SMNΔ7) that is rapidly degraded. Cis-acting elements promoting exon inclusion are indicated with green plus signs, while inhibitory elements are marked with red minus signs.