Back to article: Base editors: a powerful tool for generating animal models of human diseases
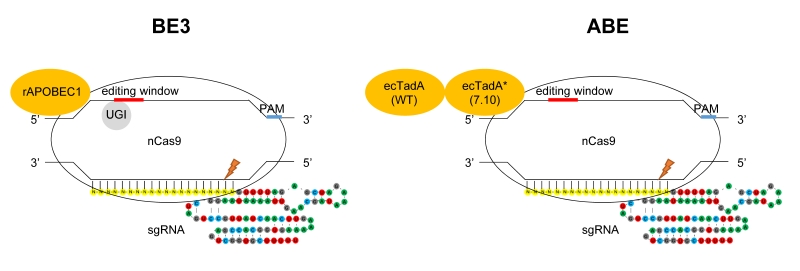
FIGURE 1: Targeted base substitutions without double strand break by cytidine- and adenine- deaminase. In these illustrations of base editing, deaminase proteins are shown in orange. The base-editing windows are highlighted in red, and the PAM sequences in blue on the DNA strand. nCas9 indicates Cas9 nickase harboring D10A mutation. Left: BE3 composed of rAPOBEC1, nCas9 and UGI (uracil glycosylase inhibitor). rAPOBEC1 indicates cytidine deaminase APOBEC1 derived from rat. Right: ABE composed of ecTadA (WT)-ecTadA* (7.10) fusion and nCas9. ecTadA (WT) indicates natural adenine deaminase Escherichia coli TadA. ecTadA* (7.10) indicates Escherichia coli TadA variant after protein evolution.