Back to article: Base editors: a powerful tool for generating animal models of human diseases
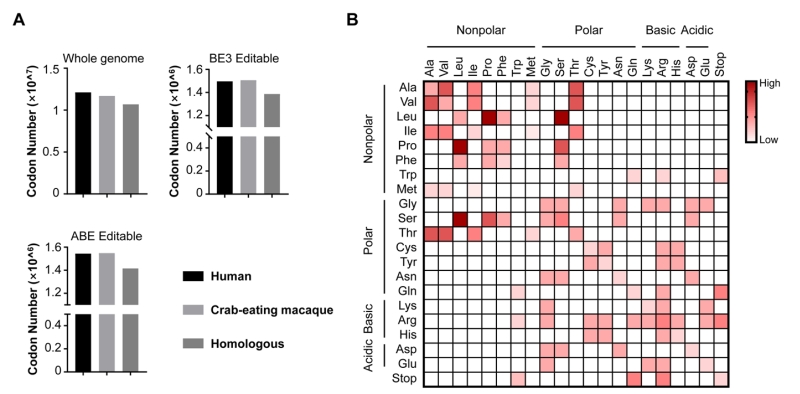
FIGURE 2: The analysis of editable codons for BEs. (A) The number of codons from NCBI database. Histograms in this panel show the number of codons of human and crab-eating macaque genomes as well as their homologous codons (Whole genome), and the number of codons from these two species editable with BE3 and ABE (BE3 Editable and ABE Editable, respectively). (B) Illustration of the repertoire of the amino acid conversions generated by BEs (BE3 and ABE). The amino acids on x axis and y axis are converted between each other. The theoretical abilities of amino acid conversions are displayed in red-white color scale.