Back to article: R-loops: formation, function, and relevance to cell stress
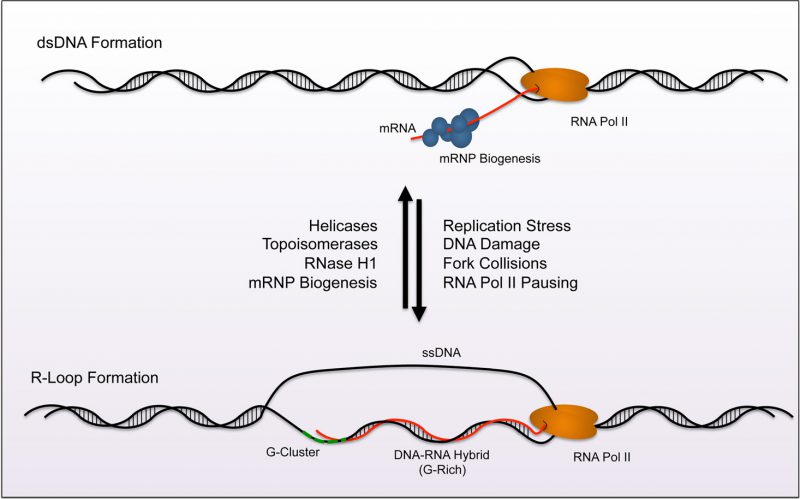
FIGURE 1: Model of R-loop homeostasis. A homeostasis exists between formation and removal of R-loops across the genome. Normally, mRNP biogenesis machinery targets nascent RNA and processes and prepares the strand for nuclear export (top). Topoisomerases and Helicases aid in preventing R-loop formation by reducing exposure times of ssDNA during transciption. RNase H removes R-loops by specifically digesting RNA in helical formation. Together these processes reduce R-loop accumulation during transcription. Factors that lead to pausing of RNA polymerase render conditions favorable for R-loop formation (replication stress, DNA damage, fork collisions, etc). R-loops nucleate from G-clusters (dashed green line). The loop then extends along GC rich sequences within the gene during elongation (bottom). The balance between removal and formation creates an equilibrium at nucleation sites across the genome.