Back to article: Control of nucleolar stress and translational reprogramming by lncRNAs
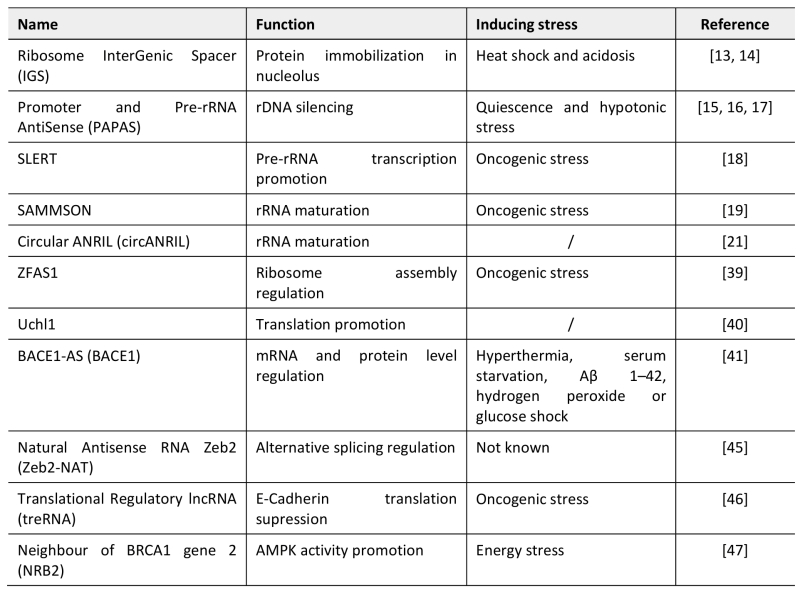
TABLE 1. LncRNAs archetypes regulating nucleolar functions and translation reprogramming during stress responses.
13. Jacob MD, Audas TE, Uniacke J, Trinkle-Mulcahy L, Lee S (2013). Environmental cues induce a long noncoding RNA-dependent remodeling of the nucleolus. Mol Biol Cell 24(18): 2943-2953. doi: 10.1091/mbc.E13-04-0223
14. Audas TE, Jacob MD, Lee S (2012). Immobilization of Proteins in the Nucleolus by Ribosomal Intergenic Spacer Noncoding RNA. Mol Cell 45(2): 147-157. doi: 10.1016/j.molcel.2011.12.012
15. Bierhoff H, Dammert MA, Brocks D, Dambacher S, Schotta G, Grummt I (2014). Quiescence-Induced LncRNAs Trigger H4K20 Trimethylation and Transcriptional Silencing. Mol Cell 54(4): 675-682. doi: 10.1016/j.molcel.2014.03.032
16. Zhao Z, Dammert MA, Grummt I, Bierhoff H (2016). lncRNA-Induced Nucleosome Repositioning Reinforces Transcriptional Repression of rRNA Genes upon Hypotonic Stress. Cell Rep 14(8): 1876-1882. doi: 10.1016/j.celrep.2016.01.073
17. Zhao Z, Dammert MA, Hoppe S, Bierhoff H, Grummt I (2016). Heat shock represses rRNA synthesis by inactivation of TIF-IA and lncRNA-dependent changes in nucleosome positioning. Nucleic Acids Res 44(17): 8144-8152. doi: 10.1093/nar/gkw496
18. Xing Y-H, Yao R-W, Zhang Y, Guo C-J, Jiang S, Xu G, Dong R, Yang L, Chen L-L (2017). SLERT Regulates DDX21 Rings Associated with Pol I Transcription. Cell 169(4): 664-678.e616. doi: 10.1016/j.cell.2017.04.011
19. Vendramin R, Verheyden Y, Ishikawa H, Goedert L, Nicolas E, Saraf K, Armaos A, Ponti RD, Izumikawa K, Mestdagh P, L.J. DL, Gian GT, Takahashi N, Marine J-C, Leucci E (2018). SAMMSON fosters cancer cell fitness by enhancing concertedly mitochondrial and cytosolic translation. Nat Struct Mol Biol 25(11):1035-1046. doi: 10.1038/s41594-018-0143-4
21. Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, Gabel G, Beutner F, Scholz M, Thiery J, Musunuru K, Krohn K (2016). Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 7: 12429. doi: 10.1038/ncomms12429
39. Hansji H, Leung EY, Baguley BC, Finlay GJ, Cameron-Smith D, Figueiredo VC, Askarian-Amiri ME (2016). ZFAS1: a long noncoding RNA associated with ribosomes in breast cancer cells. Biol Direct 11(1): 1-25. doi: 10.1186/s13062-016-0165-y
40. Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, Forrest AR, Carninci P, Biffo S, Stupka E, Gustincich S (2012). Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491(7424): 454-457. doi: 10.1038/nature11508
41. Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, Laurent Iii GS, Kenny PJ, Wahlestedt C (2008). Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of β-secretase. Nat Med 14(7): 723-730. doi: 10.1038/nm1784
45. Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG (2008). A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev 22(6): 756-769. doi: 10.1101/gad.455708
46. Gumireddy K, Li A, Yan J, Setoyama T, Johannes GJ, Orom UA, Tchou J, Liu Q, Zhang L, Speicher DW, Calin GA, Huang Q (2013). Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO J 32(20): 2672-2684. doi: 10.1038/emboj.2013.188
47. Liu X, Xiao Z-D, Han L, Zhang J, Lee S-W, Wang W, Lee H, Zhuang L, Chen J, Lin H-K, Wang J, Liang H, Gan B (2016). LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol 18(4): 431-442. doi: 10.1038/ncb3328