Back to article:
RPA and Pif1 cooperate to remove G-rich structures at both leading and lagging strand
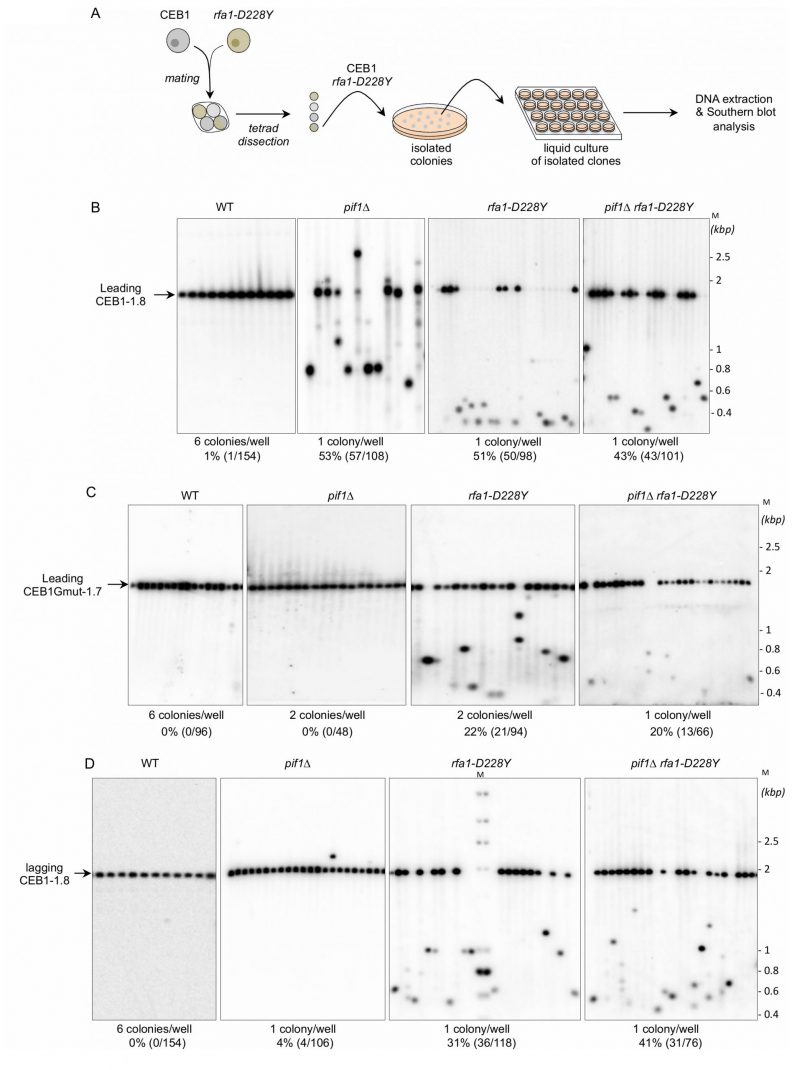
FIGURE 2: RPA is required to stabilize CEB1. (A) Experimental scheme. Yeast strains of interest are mated and the resulting diploid sporulated and dissected. After identification of spore-colonies of interest, the spore-colony is plated on media to obtain isolated colonies. Individual colonies are placed in liquid culture until stationary growth phase. Genomic DNAs are extracted and analysed by Southern blot. (B) RPA is required to stabilize CEB1 when the G4-forming strand is replicated by the leading polymerase. Genomic DNAs from yeast cells bearing the leading-CEB1 were digested by ApaI and XhoI, and southern blotted. Membranes were hybridized with the CEB1-0.6 probe. (C) Genomic DNAs from yeast cells containing the leading-CEB1Gmut-1.7 were digested by ApaI and SacII, and southern blotted. The membranes were hybridized with the CEB1Gmut-1.7 probe. (D) In contrast to Pif1, RPA is required to stabilize CEB1 when the G4-forming strand is replicated by the lagging polymerase. Genomic DNAs from yeast cells bearing the lagging-CEB1 were digested by ApaI and NcoI, and southern blotted. Membranes were hybridized with the CEB1-0.6 probe. M: ladder DNA serving as size standard (kbp). The number of colonies analysed per well, the percentage of rearrangement frequencies, and the total numbers of colonies are indicated in Table 1.