Back to article: Role of RNA Binding Proteins with prion-like domains in muscle and neuromuscular diseases
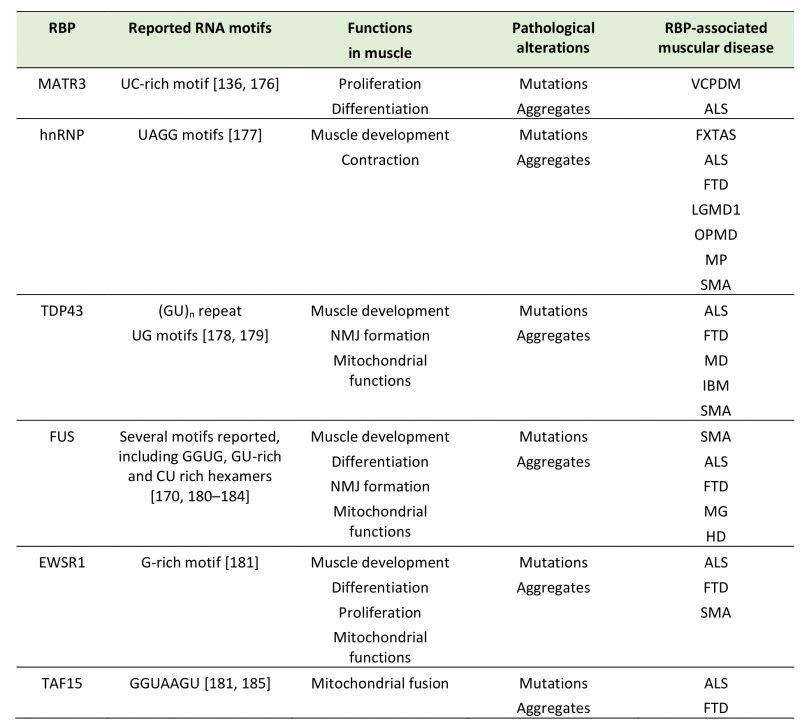
TABLE 1. Summary of selected RBP with prion like domain in neuromuscular disease.
ALS: Amyotrophic lateral sclerosis, DM: Distal myopathy, FXTAS: Fragile X-associated tremor/ataxia syndrome, HD: Huntington disease, IBM: Inclusion body myopathy, LGMD1: limb-girdle muscular dystrophy 1D, MD: Muscular dystrophy, MG: Myasthenia gravis, MP: Multisystem proteinopathy, OPMD: Oculopharyngeal muscular dystrophy, SMA: Spinal muscular atrophy, VCPDM: Vocal cord and pharyn-geal weakness with distal myopathy.
136. Coelho MB, Attig J, Bellora N, König J, Hallegger M, Kayikci M, Eyras E, Ule J, and Smith CWJ (2015). Nuclear matrix protein Matrin3 regulates alternative splicing and forms overlapping regulatory networks with PTB. EMBO J 34(5): 653–668. 10.15252/embj.201489852
170. Lagier-Tourenne C, Polymenidou M, Hutt KR, Vu AQ, Baughn M, Huelga SC, Clutario KM, Ling S-C, Liang TY, Mazur C, Wancewicz E, Kim AS, Watt A, Freier S, Hicks GG, Donohue JP, Shiue L, Bennett CF, Ravits J, Cleveland DW, and Yeo GW (2012). Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci 15(11): 1488–1497. 10.1038/nn.3230
176. Uemura Y, Oshima T, Yamamoto M, Reyes CJ, Costa Cruz PH, Shibuya T, and Kawahara Y (2017). Matrin3 binds directly to intronic pyrimidine-rich sequences and controls alternative splicing. Genes Cells 22(9): 785–798. 10.1111/gtc.12512
177. Burd CG, and Dreyfuss G (1994). Conserved structures and diversity of functions of RNA-binding proteins. Science 265(5172): 615–621. 10.1126/science.8036511
178. Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, König J, Hortobágyi T, Nishimura AL, Zupunski V, Patani R, Chandran S, Rot G, Zupan B, Shaw CE, and Ule J (2011). Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci 14(4): 452–458. 10.1038/nn.2778
179. Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling S-C, Sun E, Wancewicz E, Mazur C, Kordasiewicz H, Sedaghat Y, Donohue JP, Shiue L, Bennett CF, Yeo GW, and Cleveland DW (2011). Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 14(4): 459–468. 10.1038/nn.2779
180. Lerga A, Hallier M, Delva L, Orvain C, Gallais I, Marie J, and Moreau-Gachelin F (2001). Identification of an RNA binding specificity for the potential splicing factor TLS. J Biol Chem 276(9): 6807–6816. 10.1074/jbc.M008304200
181. Hoell JI, Larsson E, Runge S, Nusbaum JD, Duggimpudi S, Farazi TA, Hafner M, Borkhardt A, Sander C, and Tuschl T (2011). RNA targets of wild-type and mutant FET family proteins. Nat Struct Mol Biol 18(12): 1428–1431. 10.1038/nsmb.2163
182. Masuda A, Takeda J, Okuno T, Okamoto T, Ohkawara B, Ito M, Ishigaki S, Sobue G, and Ohno K (2015). Position-specific binding of FUS to nascent RNA regulates mRNA length. Genes Dev 29(10): 1045–1057. 10.1101/gad.255737.114
183. Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, and Sabatini DM (2015). Identification and characterization of essential genes in the human genome. Science 350(6264): 1096–1101. 10.1126/science.aac7041
184. Rogelj B, Easton LE, Bogu GK, Stanton LW, Rot G, Curk T, Zupan B, Sugimoto Y, Modic M, Haberman N, Tollervey J, Fujii R, Takumi T, Shaw CE, and Ule J (2012). Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain. Sci Rep 2: 603. 10.1038/srep00603
185. Kapeli K, Pratt GA, Vu AQ, Hutt KR, Martinez FJ, Sundararaman B, Batra R, Freese P, Lambert NJ, Huelga SC, Chun SJ, Liang TY, Chang J, Donohue JP, Shiue L, Zhang J, Zhu H, Cambi F, Kasarskis E, Hoon S, Jr MA, Burge CB, Ravits J, Rigo F, and Yeo GW (2016). Distinct and shared functions of ALS-associated proteins TDP-43, FUS and TAF15 revealed by multisystem analyses. Nat Commun 7(1): 1–14. 10.1038/ncomms12143