Reviews:
Cell Stress, Vol. 5, No. 8, pp. 119 - 127; doi: 10.15698/cst2021.08.254
PDLIM1: Structure, function and implication in cancer
1 Laboratory of Molecular Oncology, Frontiers Science Center for Disease-related Molecular Network, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, Chengdu 610041, China.
2 Department of Neurosurgery, West China Hospital, Sichuan University, Chengdu 610041, China.
Keywords: PDLIM1, CLP36, cytoskeleton, EMT, cancer progression.
Abbreviatons:
AA – amino acid;
ACTN – α-actinin;
ALP – actinin-associated LIM protein;
Clik1 – CLP-36 interacting kinase 1;
CLP36 – carboxyl terminal LIM domain protein of 36 kDa;
CML – chronic myelogenous leukaemia;
CRC – colorectal cancer;
DRG – dorsal root ganglion;
EMT – epithelial-mesenchymal transition;
ES – ectoplasmic specialization;
HCC – hepatocellular carcinoma;
neoHFF – neonatal human foreskin fibroblasts;
PDLIM1 – PDZ and LIM domain protein 1;
RIL – reversion-induced LIM protein.
Received originally: 23/05/2021 Received in revised form: 27/06/2021
Accepted: 28/06/2021
Published: 26/07/2021
Correspondence:
Yong Peng, Laboratory of Molecular Oncology, Frontiers Science Center for Disease-related Molecular Network, State Key Laboratory of Biother-apy, Sichuan University, Chengdu 610041, China; yongpeng@scu.edu.cn
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Jian-Kang Zhou, Xin Fan, Jian Cheng, Wenrong Liu and Yong Peng (2021). PDLIM1: Structure, function and implication in cancer. Cell Stress 5(8): 119-127. doi: 10.15698/cst2021.08.254
Abstract
PDLIM1, a member of the PDZ-LIM family, is a cytoskeletal protein and functions as a platform to form distinct protein complexes, thus participating in multiple physiological processes such as cytoskeleton regulation and synapse formation. Emerging evidence demonstrates that PDLIM1 is dysregualted in a variety of tumors and plays essential roles in tumor initiation and progression. In this review, we summarize the structure and function of PDLIM1, as well as its important roles in human cancers.
INTRODUCTION
PDLIM1 (PDZ and LIM domain protein 1), also known as CLP36, Elfin or CLIM1, is an important player in cytoskeletal organization, neuronal signaling and organ development through interacting with a variety of proteins, such as α-actinin, paladin, FHL1 and EGFR [1][2][3][4][5]. Four α-actinin genes (ACTN1-4) are embedded in human genome and their expression exhibits tissue-specific patterns. ACTN2 and ACTN3 are highly expressed in skeletal muscle and cardiac muscle sarcomere, respectively, whereas ACTN1 and ACTN4 are widely expressed in non-muscle cells [3][6][7][8][9]. PDLIM1 forms a complex with α-actinin-1/4 in colonic epithelial cells and localizes to actin stress fibers [1][6], while the interaction between PDLIM1 and α-actinin-2 was found in human myocardium [10]. What's more, in the dorsal root ganglion neurons PDLIM1 interacts with palladin to influence neurite outgrowth during sciatic nerve regeneration [2].
–
Elaborate regulation of PDLIM1 is crucial for maintaining homeostasis under physiological conditions. However, increasing evidence recently demonstrates that PDLIM1 is dysregulated in a variety of tumors, such as colorectal cancer (CRC) [11], hepatocellular carcinoma (HCC) [12], breast cancer [13], pancreatic cancer [14], glioma [15][16] and chronic myelogenous leukaemia (CML) [17]. Moreover, PDLIM1 plays important roles in cell proliferation and metastasis during tumor initiation and progression. In this review, we summarize the structure and function of PDLIM1 and outline the research progress of PDLIM1 in human cancers.
STRUCTURE OF PDLIM1 PROTEIN
PDLIM1 belongs to the actinin-associated LIM protein (ALP) subfamily of PDZ-LIM protein family [18]. The proteins of the ALP subfamily include ALP, reversion-induced LIM protein (RIL), Mystique and PDLIM1, which are characterized by a PDZ domain at the amino terminus and a LIM domain at the carboxyl terminus [18][19].
–
PDLIM1 was originally cloned from normoxic rat hepatocytes in 1995. The rat PDLIM1 is encoded by 327 amino acids (AAs) and is highly homologous to many LIM domain proteins, particularly rat RIL [20][21]. The overall homology of PDLIM1 to rat RIL was 45.1%, with 62% homology at the N-terminus (AA 1-89) and 50% homology at the C-terminus (AA 192-327). Subsequently, Kotaka et al. characterized the human PDLIM1 cDNA clone [22]. Since the ORF of this cDNA encodes a 36-kDa carboxyl terminal LIM domain protein with a PDZ domain at the amino terminal, which shares high homology to rat CLP36 (36 kDa carboxy-terminal LIM domain protein), the encoded protein was named human 36 kDa carboxy-terminal LIM domain protein (hCLIM1) [10][22]. Human PDLIM1 is composed of 329 AAs and represents high conservation and homology in structure among species (Figure 1). The identity of its PDZ domain to human ALP is 55% and that of human RIL is 66%, while the LIM domain identity was 67% to ALP and 57% to RIL, respectively [1]. In view of the high sequence homology between PDLIM1 and other proteins in the ALP subfamily, it is not surprising that the function of PDLIM1 in cells can be partially rescued by proteins such as RIL [23], whereas in some cases it cannot be compensated [13]. Therefore, the specific mechanisms of PDLIM1 and other ALP family proteins are worthy of in-depth study.
–
–
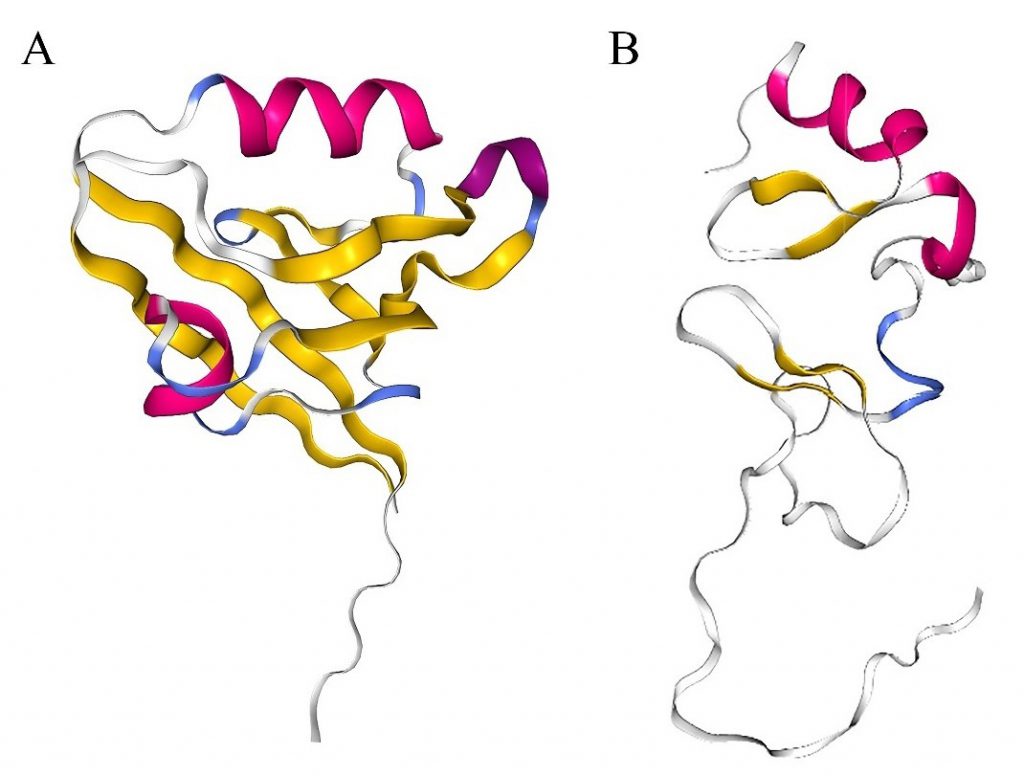
Since the protein structure of PDLIM1 has not been fully analyzed, expounding its two domains, PDZ and LIM, may be helpful to understand the role of PDLIM1. The PDZ domain (AA 3-85 of PDLIM1, PDB ID: 2PKT), whose name is derived from PSD-95, DLG and ZO-1 proteins that were first discovered to contain this domain, consists of antiparallel β chains and α helices [24][25][26] (Figure 2A). The PDZ domain is highly conserved and widely present in the proteins of bacteria, fruit flies, plants and animals. As an important domain that mediates protein interactions, it provides protein binding sites and can mediate the formation of multiple protein complexes, including membrane-associated proteins, cytoplasmic signaling proteins, and cytoskeletal proteins [27][28]. The proteins that have been reported to bind to the PDZ domain of PDLIM1 include α-actinin, p75NTR and β-catenin/E-cadherin complex [6][11][13][15]. Another domain of PDLIM1, the LIM domain (AA 258-317 of PDLIM1, PDB ID: 1X62), was named after its first recognition in Lin11, Isl-1 and Mec-3 [29][30][31]. The LIM domain consists of two zinc finger domains exhibiting a consistent Cys-rich sequence (Cys-X2-Cys-X17±1-His-X2-Cys)-X2-(Cys-X2-Cys-X17±1-Cys-X2-His/Asp/Cys) (X represents any AA) (Figure 2B) [32][33][34]. The LIM domain is responsible for the interaction between PDLIM1 and proteins such as kinases and actin cytoskeletal components [10][35]. Generally, through PDZ and LIM domains, PDLIM1 interacts with α-actinin, palladin, and kinases, as well as serves as a scaffold to promote the formation of protein complexes, thereby regulating signal pathways and affecting cell activity [36][37].
–
BIOLOGICAL FUNCTION OF PDLIM1
PDLIM1 is widely distributed in heart, lung, liver, and spleen tissues [18][38]. By interacting with various proteins in different tissues, PDLIM1 participates in the regulation of multiple signal transduction pathways and exhibits tissue-specific functions.
–
As a cytoskeleton-associated protein, PDLIM1 is involved in the regulation of actin cytoskeleton organization. The dynamic regulation of the assembly and disassembly of stress fibers is essential for cytoskeleton-dependent functions such as morphological changes and migration [39][40]. Previous studies have shown that inhibition of PDLIM1 impairs the assembly of focal adhesion and the formation of stress fibers in BeWo cells, and these effects can be reversed by expressing exogenous PDLIM1 (CLP36), rather than by expressing its mutant form of lacking PDZ or LIM domains, indicating the important role of PDZ and LIM domains in maintaining cytoskeleton-dependent functions [41]. Moreover, PDLIM1-deficienct fibroblasts exhibit a reduction in stress fiber formation and a significant increase in directional and random migration, and these abnormalities caused by PDLIM1 deletions can be compensated by RIL protein [23]. In addition to affecting the formation and dynamics of stress fibers, PDLIM1 can also act as an adapter molecule that recruits signal molecules to stress fibers. For example, Vallenius et al. confirmed the LIM domain-dependent interaction between PDLIM1 and CLP-36 interacting kinase 1 (Clik1) by yeast two-hybrid analysis. This interaction with PDLIM1 makes Clik1, which is originally located in the nucleus, dramatically relocate to actin stress fibers [35]. However, whether this relocation leads to changes in the function of Clik1 remains to be explored. Furthermore, a recent study showed that PDLIM1 resides in the actin-rich structure induced by invasive and adherent bacterial pathogens, such as Listeria monocytogenes and enteropathogenic Escherichia coli (EPEC), but the specific mechanism still needs further study [42].
–
The correct assembly of the cytoskeleton regulated by PDLIM1 is essential for the normal function of Sertoli cells and the ordered spermatid differentiation during spermiogenesis [43][44]. Ectoplasmic specialization (ES) is a testis-unique anchoring junction and essential for Sertoli-germ cell communication and successful spermatogenesis [45]. Under normal circumstances, redundant PDLIM1 is degraded by the autophagic lysosomal pathway in Sertoli cells to promote proper assembly of ES and maintain an appropriate cytoskeletal network during sperm formation. However, when autophagy is destroyed by knocking out autophagy-related proteins ATG7 or ATG5, PDLIM1 is accumulated in the cytoplasm of Sertoli cells and disrupts the F-actin hoops of apical ES and related microtubule-based structures in the seminiferous epithelium, leading to disordered cytoskeleton structures and broken ES assembly, thus affecting Sertoli-germ cell communication [43]. Similarly, knockout of Atg7 in spermatids blocks autophagy and upregulates PDLIM1 in the cytoplasm, resulting in the formation of stress fibers-like structures and the disruption of spermatozoa flagella assembly and spermatid differentiation, eventually leading to sterility [44].
–
It is reported that PDLIM1 is expressed in the sensory ganglia of adult rats, but not in the central nervous system, and its expression is upregulated in peripheral sensory neurons and motor neurons after sciatic nerve transection, suggesting that PDLIM1 may play a certain role in the regulation of neurite growth [46]. In primary dorsal root ganglion (DRG) neurons and undifferentiated PC12 cells, PDLIM1 is distributed in the cell body and neurites and is highly enriched in the growth cone. Functionally, overexpression of the PDLIM1 PDZ domain inhibits the outgrowth of neurite. Instead, knockdown of PDLIM1 in PC12 cells with shRNA altered cell morphology and activated growth cone movements, resulting in increased length and number of neurites. Likewise, inhibition of PDLIM1 in primary DRG neurons increased the growth rate of neurite cells [46]. These results demonstrate that PDLIM1 plays an important role in controlling neurite outgrowth.
–
Furthermore, PDLIM1 is also involved in the regulation of signaling pathways. The transcription factor NF-κB, a class of dimer proteins consisting of p65 (RelA), NF-κB1 (p50/p105), NF-κB2 (p52/p100), c-Rel and RelB, is usually inactivated through binding to its inhibitor protein IκB [47]. When stimulated by Toll-like receptors (TLR) signaling, IκB can be phosphorylated by the activated IKK complex and is eventually degraded by the ubiquitin ligase machinery. Thus, the freed NF-κB dimer is translocated into the nucleus, where it initiates the transcription of target genes related to the inflammatory reaction [48][49]. PDLIM1 was reported to be an important factor in modulating p65 nuclear translocation, thereby participating in the regulation of the NF-κB pathway. In dendritic cells, p65 was shown to bind to PDLIM1 and thus be retained in the cytoplasm, thereby reducing its nuclear translocation and NF-κB-mediated inflammatory signaling. Notably, the inhibition of NF-κB signaling by PDLIM1 is independent of IκBα, but dependent on α-actinin-4, the binding protein of PDLIM1, because knocking down α-actinin-4 reversed the inhibition of p65 nuclear translocation mediated by PDLIM1. Besides, deficiency of PDLIM1 in mice leads to the elevated levels of nuclear p65 protein, and thus promotes the production of proinflammatory cytokines and chemokines (such as IL-6, IL-12, TNFα, IL-18, CXCL2 and CXCL10), indicating that PDLIM1 negatively regulates NF-κB-mediated inflammation in innate immune response [50]. Therefore, PDLIM1-mediated inhibition of the NF-κB pathway may serve as a new strategy for the treatment of inflammatory diseases.
PDLIM1 AND CANCER PROGRESSION
PDLIM1 plays an important role in maintaining certain homeostasis of cells. The dysregulation of PDLIM1 usually manifests as its abnormal expression, which affects the interaction with its binding proteins and the related signal pathways (such as NF-κB, Wnt/β-catenin and Hippo), leading to the occurrence and development of many diseases including cancer.
–
PDLIMI and CRC
CRC is the third most common cancer worldwide, accounting for approximately 10% of all new cases and deaths each year [51]. The major cause of death in CRC patients is metastasis, which can be promoted by epithelial-mesenchymal transition (EMT) [52][53]. EMT is a biological process in which epithelial cells are transformed into mesenchymal cells under specific conditions, characterized by the down-regulation of epithelial factors (E-cadherin, claudins) and activation of mesenchymal factors (N-cadherin, vimentin) [54]. In CRC, deregulation of the canonical Wnt/β-catenin signaling pathway increases SNAIL, leading to the reduction of E-cadherin and thus promoting EMT [55]. Moreover, β-catenin and E-cadherin can form an E-cadherin/β-catenin complex, which can maintain tight junctions between cells and prevent cell invasion and metastasis. Factors such as TGF-β can dissociate and translocate β-catenin from the E-cadherin/β-catenin to the nucleus, thereby activating the Wnt pathway and inducing EMT [56][57][58].
–
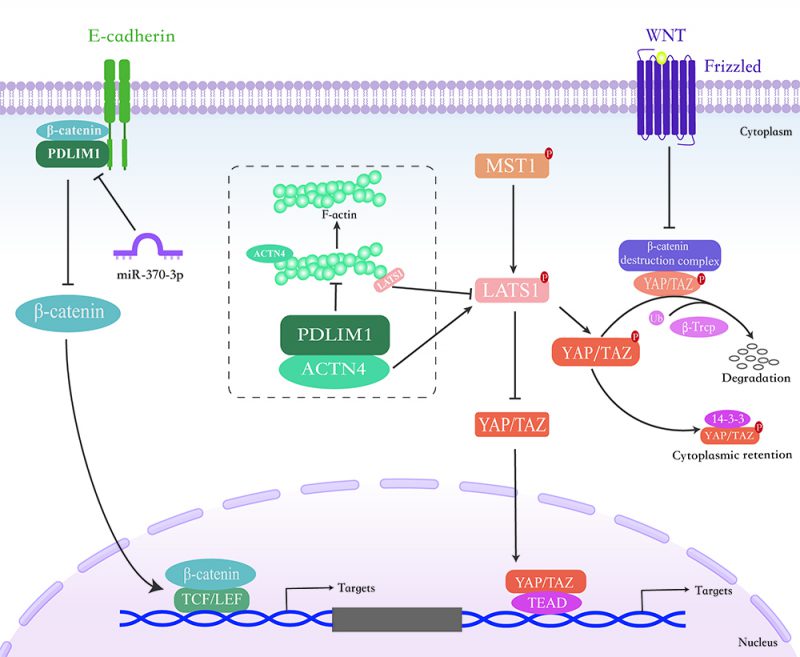
Chen et al. found that PDLIM1 was down-regulated in CRC tissues compared to peritumoral tissues due to the hypermethylation of the PDLIM1 gene promoter in CRC tissues. Importantly, loss of PDLIM1 promotes the invasion and metastasis of CRC cells in vitro and in vivo. By contrast, enhanced PDLIM1 inhibits CRC cell invasion and metastasis. Moreover, the deletion of PDLIM1 down-regulates the expression of epithelial factors and promotes the expression of mesenchymal factors, indicating that PDLIM1 promotes the invasion and metastasis of CRC cells by affecting the EMT process. Mechanistically, down-regulation of PDLIM1 in CRC cells reduces the binding of β-catenin and E-cadherin, leading to the β-catenin nuclear translocation to initiate EMT and metastatic process in CRC cells, suggesting that PDLIM1 is involved in the regulation of cell invasion and migration and exerts a tumor suppressor function in CRC. Furthermore, repression of PDLIM1 in CRC tissues predicts poor survival in CRC patients [11]. Taken together, PDLIM1 can interact with E-cadherin/β-catenin complex, thereby suppressing the transcriptional activity of β-catenin and inhibiting the occurrence of EMT in CRC (Figure 3). Therefore, PDLIM1 may serve potentially as a marker of tumor aggressiveness and as a predictor of survival in CRC patients.
–
–
Intriguingly, Rai et al. reported that PDLIM1 in neonatal human foreskin fibroblasts (neoHFFs) can act as a pro-invasive regulator of membrane protrusion. PDLIM1 is upregulated in neoHFFs activated by late-stage colorectal cancer-exosomes (exosomes derived from SW620 CRC cells), allowing cells to acquire the ability to invade through the extracellular matrix [59]. Accordingly, the function of PDLIM1 in CRC may be highly context-dependent, which provides more perspectives for the study of PDLIM1.
–
PDLIM1 and HCC
The Hippo pathway is an evolutionarily conserved tumor-suppressor signaling pathway, which plays an essential role in a variety of tumors including HCC [60][61]. The core components of the Hippo pathway include MST1/2, LATS1/2, YAP, and downstream target genes. When Hippo signaling is on, activated MST1/2 kinases can phosphorylate and activate its substrate LATS1/2, which in turn phosphorylates the effector YAP. The phosphorylated YAP are retained in the cytoplasm due to the interaction with the 14-3-3 protein and further binds to the β-Catenin destruction complex to trigger β-TrCP-mediated degradation. However, when Hippo signaling is off, YAP is stabilized and translocated to the nucleus, and combines with transcriptional activators to initiate transcription of downstream genes such as CTGF, CYR61, thereby promoting tumor progression [62].
–
Our group reported that PDLIM1 expression is significantly decreased in metastatic HCC tissues compared to non-metastatic tumors, suggesting that PDLIM1 may play an inhibitory role during HCC metastasis [12]. Functional studies showed that knockdown of PDLIM1 in HCC cells can induce EMT and promote metastasis in vitro and in vivo, while overexpression of PDLIM1 shows the opposite phenotype. To examine the associated protein of PDLIM1 in HCC, we performed immunoprecipitation and mass spectrometric analysis, and identified that PDLIM1 interacts with the cytoskeleton cross-linking protein ACTN4. Specifically, PDLIM1 competitively binds to ACTN4 through Asn145 (N145), thereby weakening the interaction between ACTN4 and F-actin and preventing the overgrowth of F-actin. Therefore, loss of PDLIM1 in HCC cells leads to the formation of excessive F-actin, which further induces LATS1 dephosphorylation, inactivates the Hippo pathway and promotes HCC metastasis (Figure 3). Furthermore, low expression of PDLIM1 is associated with poor prognosis in HCC patients [12]. Therefore, our findings dissect the role and the underlying mechanism of PDLIM1 in HCC metastasis, and also highlight its prognostic potential in HCC.
–
PDLIM1 and breast cancer
Breast cancer accounts for 30% of female malignancies and has a mortality rate of 15% worldwide [51]. Pitteri et al. reported that PDLIM1 expression is increased during breast cancer progression, suggesting that PDLIM1 may promote breast cancer development [63]. Liu et al. further confirmed that PDLIM1 is a pro-metastatic factor in breast cancer and enhances the invasion and metastasis of breast cancer cells through α-actinin-Cdc42 signaling pathway. PDLIM1 knockdown by RNAi in MDA-MB-231 cells significantly represses metastasis in vitro and in vivo, while overexpression of PDLIM1 exhibits opposite phenotypes. Importantly, PDLIM1 combines with α-actinin-4 to form a PDLIM1-α-actinin complex, thereby promoting the activation of Cdc42, which is a key factor in regulating cell polarity and migration of breast cancer cells [13][64]. Notably, the PDZ domain is responsible for the binding of PDLIM1 to α-actinin in breast cancer cells. Disruption of this interaction by deleting the PDZ domain eliminates PDLIM1-mediated cell migration, further demonstrating the essential role of PDLIM1-α-actinin complex in breast cancer migration and invasion. Based on these findings, targeting PDLIM1-α-actinin-Cdc42 axis may have a potential for inhibiting breast cancer metastasis and is worthy of further investigation.
–
PDLIM1 and pancreatic cancer
Pancreatic cancer is a highly malignant digestive tract tumor, with a five-year survival rate of only 9% due to the difficulty to diagnose and treat [51]. The identification of tumor antigens provides the possibility for early diagnosis and targeted therapy of patients with pancreatic cancer. Dr. Hong analyzed the sera of 36 patients with pancreatic cancer and 68 sera from healthy donors and patients with other cancers to identify proteins that specifically induce humoral responses in pancreatic cancer patients. Proteins extracted from the human pancreatic cancer BxPC-3 cells were separated by 2-Dimensional polyacrylamide gel electrophoresis (2-D PAGE), and then transferred to the PVDF membrane. The membranes were incubated with the collected sera individually to detect the antibodies against BxPC3 proteins, and a protein of approximately 36kDa showed reactivity with sera from 38.9% patients of pancreatic cancer and 4.4% of the other two groups, suggesting its pancreatic cancer specificity. Mass spectrometric analysis was performed to confirm that the protein was PDLIM1 [14]. The pancreas/ampullary adenocarcinoma tissue array further proved the immunoreactivity of PDLIM1 against pancreatic cancer. In addition, PDLIM1 RNA levels were significantly higher in pancreatic cancer cell lines compared to colon, lung, and ovarian cancer cell lines, and PDLIM1 was located in both cell membrane and cytoplasm, suggesting that PDLIM1 may be a tumor autoantigen in pancreatic cancer. However, the role of PDLIM1 in pancreatic cancer still needs further research, which may help to explore the potential of PDLIM1 as an early diagnostic marker or drug target for pancreatic cancer.
–
PDLIM1 and glioma
The p75 neurotrophin receptor (p75NTR, also known as CD271) is a key molecule that drives tumorigenesis, invasion and metastasis in glioma [65][66]. Ahn et al. demonstrated PKA inhibition reduced the phosphorylation of p75NTR at S303 and thus attenuated p75NTR-mediated glioma invasion, indicating that PKA-induced p75NTR phosphorylation at S303 is required for glioma invasion. Intriguingly, a novel phosphorylation at S425 that located within the C-terminal PDZ binding motif of p75NTR was detected by nano-liquid chromatography–tandem mass spectrometry in cells expressing p75NTR. Further experiments proved that the deletion or mutation of the PDZ binding motif resulted in the elimination of p75NTR-mediated glioma invasion, suggesting that p75NTR and its interacting PDZ domain-containing protein may work together to regulate glioma invasion. Therefore, the p75NTR-interacting protein containing the PDZ domain was identified by mass spectrometry, and it was found that PDLIM1 may be a potential binding protein of p75NTR. Immunoprecipitation and in vitro pull-down experiments further demonstrated the direct interaction between PDLIM1 and the C-terminal cytoplasmic domain of p75NTR, and this interaction was reduced when S425 of p75NTR was phosphorylated, proving that S425 is a key residue to mediate the interaction between PDLIM1 and p75NTR. Moreover, knockdown of PDLIM1 by shRNA abolished p75NTR-mediated glioma invasion in vitro and in vivo[15][16]. Overall, the interaction of p75NTR and PDLIM1, which depends on the unphosphorylated state of S425, mediates the invasion of glioma. Consequently, disruption of the interaction between the PDZ domain of PDLIM1 and p75NTR may provide potential therapeutic strategies for patients with glioma.
–
PDLIM1 and CML
CML is a hematopoietic malignancy derived from the abnormal proliferation of bone marrow hematopoietic stem cells, and accounts for 15% of new cases of leukemias in adults [67][68]. MicroRNA, a class of small non-coding RNA that play essential roles in CML progression [69][70][71]. Recently, Li et al. reported that miR-370-3p can suppress the proliferation and induce apoptosis of CML cells by targeting PDLIM1 [17]. The expression of miR-370-3p was found significantly reduced in the peripheral blood mononuclear cells (PBMC) from CML patients. Functional studies indicate that miR-370-3p inhibits proliferation and promotes apoptosis of CML cells. Further research revealed that miR-370-3p can directly bind to the 3'-untranslated region of PDLIM1, proving that PDLIM1 is a target gene of miR-370-3p. In view of the role in promoting proliferation and suppressing apoptosis, PDLIM1 was found to be an oncogene in CML, and the overexpression of PDLIM1 weakened the tumor suppressive role of miR-370-3p in CML, indicating that miR-370-3p functions partially depends on the repression of PDLIM1. Moreover, the author proved that miR-370-3p inhibits Wnt/β-catenin signaling by targeting PDLIM1, thereby suppressing CML cell proliferation and inducing apoptosis. Collectively, these results demonstrate the vital role of miR-370-3p-PDLIM1-Wnt/β-catenin signaling axis in the progression of CML (Figure 3). Therefore, the development of drugs targeting the miR-370-3p-PDLIM1-Wnt/β-catenin signaling axis, combined with other strategies, may be helpful to improve the clinical treatment of CML.
CONCLUDING REMARKS
PDLIM1 is a highly conserved cytoskeleton protein that belongs to the PDZ and LIM protein family. Under normal circumstances, PDLIM1 participates in maintaining homeostasis within cells. In cancer, however, the dysregulation of PDLIM1 can lead to abnormalities in a series of signaling pathways. Through interacting with actin filaments and other proteins, PDLIM1 can regulate the proliferation, metastasis and survival of malignant cells, and ultimately affect the progression of cancers. Therefore, more research on the function of PDLIM1 is helpful for the diagnosis and treatment of tumors in the clinic.
–
Tumor cells acquire migration and invasion capabilities through EMT, which refers to dynamic reorganization of the cytoskeleton. As a cytoskeleton protein, PDLIM1 is involved in the EMT process in a variety of tumors. Interestingly, PDLIM1 displays remarkable tissue-specific roles across cancers. It can inhibit the metastasis of CRC [11] and HCC [12], but promote the development of various cancers such as breast cancer [13] and CML [17]. It is well known that the process of tumorigenesis and development involves the combined action of multiple genes and cellular signaling pathways. Therefore, it seems reasonable to speculate that the tissue-specific characteristics of PDLIM1 may depend on the difference in its interacting proteins and regulatory signaling pathways in different cancer types. Although progress has been made in characterizing PDLIM1, the cellular and molecular mechanisms of PDLIM1 and the corresponding downstream signaling pathways in cancer still need further research, which may help deepen the understanding of cancer progression and provide new strategies for the optimization of targeted therapy or combined therapy in the clinic.
REFERENCES
- Bauer K, Kratzer M, Otte M, de Quintana KL, Hagmann J, Arnold GJ, Eckerskorn C, Lottspeich F, Siess W (2000). Human CLP36, a PDZ-domain and LIM-domain protein, binds to alpha-actinin-1 and associates with actin filaments and stress fibers in activated platelets and endothelial cells. Blood 96(13): 4236-4245. 10.1016/S0002-9394(99)00202-0
- Hasegawa T, Ohno K, Funahashi S, Miyazaki K, Nagano A, Sato K (2010). CLP36 interacts with palladin in dorsal root ganglion neurons. Neurosci Lett 476(2): 53-57. 10.1016/j.neulet.2010.03.081
- Vallenius T, Scharm B, Vesikansa A, Luukko K, Schäfer R, Mäkelä TP (2004). The PDZ-LIM protein RIL modulates actin stress fiber turnover and enhances the association of alpha-actinin with F-actin. Exp Cell Res 293(1): 117-128. 10.1016/j.yexcr.2003.09.004
- Sharma P, Shathasivam T, Ignatchenko V, Kislinger T, Gramolini AO (2011). Identification of an FHL1 protein complex containing ACTN1, ACTN4, and PDLIM1 using affinity purifications and MS-based protein-protein interaction analysis. Mol Biosyst 7(4):1185-1196. 10.1039/c0mb00235f
- Naegle KM, White FM, Lauffenburger DA, Yaffe MB (2012). Robust co-regulation of tyrosine phosphorylation sites on proteins reveals novel protein interactions. Mol Biosyst 8(10):2771-2782. 10.1039/c2mb25200g
- Vallenius T, Luukko K, Makela TP (2000). CLP-36 PDZ-LIM protein associates with nonmuscle alpha-actinin-1 and alpha-actinin-4. J Biol Chem 275(15): 11100-11105. 10.1074/jbc.275.15.11100
- Beggs AH, Byers TJ, Knoll JH, Boyce FM, Bruns GA, Kunkel LM (1992). Cloning and characterization of two human skeletal muscle alpha-actinin genes located on chromosomes 1 and 11. J Biol Chem 267(13): 9281-9288. 10.1016/s0021-9258(19)50420-3
- Honda K, Yamada T, Endo R, Ino Y, Gotoh M, Tsuda H, Yamada Y, Chiba H, Hirohashi S (1998). Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion. J Cell Biol 140(6): 1383-1393. 10.1083/jcb.140.6.1383
- Otey CA, Carpen O (2004). Alpha-actinin revisited: a fresh look at an old player. Cell Motil Cytoskeleton 58(2): 104-111. 10.1002/cm.20007
- Kotaka M, Kostin S, Ngai SM, Chan K, Lau Y, Lee SM, Li Hy, Ng EK, Schaper J, Tsui SK, Fung Kp, Lee Cy, Waye MM (2000). Interaction of hCLIM1, an enigma family protein, with alpha-actinin 2. J Cell Biochem 78: 558-565. 10.1002/1097-4644(20000915)78:4<558::aid-jcb5>3.0.co;2-i
- Chen HN, Yuan K, Xie N, Wang K, Huang Z, Chen Y, Dou Q, Wu M, Nice EC, Zhou ZG, Huang C (2016). PDLIM1 stabilizes the E-Cadherin/β-Catenin complex to prevent epithelial-mesenchymal transition and metastatic potential of colorectal cancer cells. Cancer Res 76(5): 1122-1134. 10.1158/0008-5472.CAN-15-1962
- Huang Z, Zhou JK, Wang K, Chen H, Qin S, Liu J, Luo M, Chen Y, Jiang J, Zhou L, Zhu L, He J, Li J, Pu W, Gong Y, Li J, Ye Q, Dong D, Hu H, Zhou Z, Dai L, Huang C, Wei X, Peng Y (2019). PDLIM1 inhibits tumor metastasis through activating Hippo signaling in hepatocellular carcinoma. Hepatology 71(5): 1643-1659. 10.1002/hep.30930
- Liu Z, Zhan Y, Tu Y, Chen K, Liu Z, Wu C (2015). PDZ and LIM domain protein 1 (PDLIM1)/CLP36 promotes breast cancer cell migration, invasion and metastasis through interaction with α-actinin. Oncogene 34(10): 1300-1311. 10.1038/onc.2014.64
- Hong SH (2005). Identification of CLP36 as a tumor antigen that induces an antibody response in pancreatic cancer. Cancer Res Treat 37(1): 71-77. 10.4143/crt.2005.37.1.71
- Ahn BY, Saldanha-Gama RF, Rahn JJ, Hao X, Zhang J, Dang NH, Alshehri M, Robbins SM, Senger DL. (2016). Glioma invasion mediated by the p75 neurotrophin receptor (p75(NTR)/CD271) requires regulated interaction with PDLIM1. Oncogene 35(11): 1411–1422. 10.1038/onc.2015.199
- Alshehri MM, Robbins SM, Senger DL (2017). The role of neurotrophin signaling in gliomagenesis: a focus on the p75 neurotrophin receptor (p75NTR/CD271). Vitam Horm 104:367-404. 10.1016/bs.vh.2016.11.001
- Li LM, Luo FJ, Song X (2020). MicroRNA-370-3p inhibits cell proliferation and induces chronic myelogenous leukaemia cell apoptosis by suppressing PDLIM1/Wnt/β-catenin signalling. Neoplasma 67(3): 509-518. 10.4149/neo_2020_190612N506
- Zheng M, Cheng H, Banerjee I, Chen J (2010). ALP/Enigma PDZ–LIM domain proteins in the heart. J Mol Cell Biol 2(2): 96-102. 10.1093/jmcb/mjp038
- te Velthuis AJ, Bagowski CP (2007). PDZ and LIM domain-encoding genes: molecular interactions and their role in development. Scientific World Journal 7: 1470–1492. 10.1100/tsw.2007.232
- Wang H, Harrison-Shostak DC, Lemasters JJ, Herman B (1995). Cloning of a rat cDNA encoding a novel LIM domain protein with high homology to rat RIL. Gene 165(2): 267-271. 10.1016/0378-1119(95)00542-e
- Kiess M, Scharm B, Aguzzi A, Hajnal A, Klemenz R, Schwarte-Waldhoff I, Schafer R (1995). Expression of ril, a novel LIM domain gene, is down-regulated in HRAS-transformed cells and restored in phenotypic revertants. Oncogene 10(1): 61-68. 7824279
- Kotaka M, Ngai SM, Garcia-Barcelo M, Tsui SK, Fung KP, Lee CY, Waye MM (1999). Characterization ft he human 36-kDa carboxyl terminal LIM domain protein (hCLIM1). J Cell Biochem 72(2): 279-285. 10.1002/(sici)1097-4644(19990201)72:2<279::aid-jcb12>3.0.co;2-7
- Miyazaki K, Ohno K, Tamura N, Sasaki T, Sato K (2012). CLP36 and RIL recruit α-actinin-1 to stress fibers and differentially regulate stress fiber dynamics in F2408 fibroblasts. Exp Cell Res 318(14): 1716-1725. 10.1016/j.yexcr.2012.05.006
- Liu X, Fuentes EJ (2019). Emerging themes in PDZ domain signaling: structure, function, and inhibition. Int Rev Cell Mol Biol 343: 129-218. 10.1016/bs.ircmb.2018.05.013
- Guo L, Ji C, Gu S, Ying K, Cheng H, Ni X, Liu J, Xie Y, Mao Y (2003). Molecular cloning and characterization of a novel human kinase gene, PDIK1L. J Genet 82(1-2): 27-32. 10.1007/bf02715878
- Harris BZ, Lim WA (2001). Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci 114(Pt 18): 3219-3231. 10.1080/15216540152846064
- Fanning AS, Anderson JM (1996). Protein-protein interactions: PDZ domain networks. Curr Biol 6(11): 1385–1388. 10.1016/s0960-9822(96)00737-3
- Zhang M, Wang W (2003). Organization of signaling complexes by PDZ-Domain scaffold proteins. Acc Chem Res 36(7): 530-538. 10.1021/ar020210b
- Freyd G, Kim SK, Horvitz HR (1990). Novel cysteine-rich motif and homeodomain in the product ft he Caenorhabditis eleguns cell lin-eage gene lin-11. Nature 344(6269): 876-879. 10.1038/344876a0
- Karlsson O, Thor S, Norberg T, Ohlsson H, Edlund T (1990). Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo-and a cys-his domain. Nature 344(6269): 879-882. 10.1038/344879a0
- Way JC, Chalfie M (1988). Mec-3, a homeobox-containing gene that specifies differentiation ft he touch receptor neurons in C. elegans. Cell 54(1): 5-16. 10.1016/0092-8674(88)90174-2
- Sanchez-Garcia I, Rabbitts TH (1994). The LIM domain: a new structural motif found in zinc-finger-like proteins. Trends Genet 10(9): 315-320. 10.1016/0168-9525(94)90034-5
- Dawid IB, Toyama R, Taira M (1995). LIM domain proteins. C R Acad Sci III 318(3): 295–306. 7788499
- Sikorska M, Krężel A, Otlewski J (2012). Femtomolar Zn2+ affinity of LIM domain of PDLIM1 protein uncovers crucial contribution of protein-protein interactions to protein stability. J Inorg Biochem 115:28-35. 10.1016/j.jinorgbio.2012.05.009
- Vallenius T, Mäkelä TP (2002). Clik1: a novel kinase targeted to actin stress fibers by the CLP-36 PDZ-LIM protein. J Cell Sci 115(Pt 10): 2067-2073. 10.1242/jcs.115.10.2067
- Krcmery J, Camarata T, Kulisz A, Simon HG (2010). Nucleocytoplasmic functions ft he PDZ-LIM protein family: new insights into organ development. Bioessays 32(2): 100-108. 10.1002/bies.200900148
- Dawid IB, Breen JJ, Toyama R (1998). LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet 14(4): 156-162. 10.1016/S0168-9525(98)01424-3
- Kotaka M, Lau YM, Cheung KK, Lee SM, Li HY, Chan WY, Fung KP, Lee CY, Waye MM, Tsui SK (2001). Elfin is expressed during early heart development. J Cell Biochem 83(3): 463-472. 10.1002/jcb.1244
- Tojkander S, Gateva G, Lappalainen P (2012). Actin stress fibers—assembly, dynamics and biological roles. J Cell Sci 125(Pt 8): 1855-1864. 10.1242/jcs.098087
- Vallenius T (2013). Actin stress fibre subtypes in mesenchymal-migrating cells. Open Biol 3(6): 130001. 10.1098/rsob.130001
- Tamura N, Ohno K, Katayama T, Kanayama N, Sato K (2007). The PDZ-LIM protein CLP36 is required for actin stress fiber formation and focal adhesion assembly in BeWo cells. Biochem Biophys Res Commun 364(3): 589-594. 10.1016/j.bbrc.2007.10.064
- Dhanda AS, Yang D, Kooner A, Guttman JA (2021). Distribution of PDLIM1 at actin-rich structures generated by invasive and adherent bacterial pathogens. Anat Rec 304(5): 919-938. 10.1002/ar.24523
- Liu C, Wang H, Shang Y, Liu W, Song Z, Zhao H, Wang L, Jia P, Gao F, Xu Z, Yang L, Gao F, Li W (2016). Autophagy is required for ectoplasmic specialization assembly in sertoli cells. Autophagy 12(5): 19. 10.1080/15548627.2016.1159377
- Shang Y, Wang H, Jia P, Zhao H, Liu C, Liu W, Song Z, Xu Z, Yang L, Wang Y, Li W (2016). Autophagy regulates spermatid differentiation via degradation of PDLIM1. Autophagy 12(9): 1575-1592. 10.1080/15548627.2016.1192750
- Mruk DD, Cheng CY (2004). Sertoli-Sertoli and Sertoli-germ cell interactions and their significance in germ cell movement in the seminiferous epithelium during spermatogenesis. Endocr Rev 25(5): 747-806. 10.1210/er.2003-0022
- Ohno K, Kato H, Funahashi S, Hasegawa T, Sato K (2009). Characterization of CLP36/Elfin/PDLIM1 in the nervous system. J Neurochem 111(3): 790-800. 10.1111/j.1471-4159.2009.06370.x
- Taniguchi K, Karin M (2018). NF-κB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol 18(5): 309-324. 10.1038/nri.2017.142
- Hayden MS, Ghosh S (2004). Signaling to NF-κB. Genes 18(18): 2195-2224. 10.1101/gad.1228704
- Takeuchi O, Akira S (2010). Pattern recognition receptors and inflammation. Cell 140(6): 805-820. 10.1016/j.cell.2010.01.022
- Ono R, Kaisho T, Tanaka T (2015). PDLIM1 inhibits NF-κB-mediated inflammatory signaling by sequestering the p65 subunit of NF-κB in the cytoplasm. Sci Rep 5: 18327. 10.1038/srep18327
- Siegel RL, Miller KD, Jemal A (2020). Cancer statistics, 2020. CA Cancer J Clin 70(1): 7-30. 10.3322/caac.21590
- Wei Q, Ye Z, Zhong X, Li L, Wang C, Myers RE, Palazzo JP, Fortuna D, Yan A, Waldman SA, Chen X, Posey JA, Basu-Mallick A, Jiang BH, Hou L, Shu J, Sun Y, Xing J, Li B, Yang H (2017). Multiregion whole-exome sequencing of matched primary and metastatic tumors revealed genomic heterogeneity and suggested polyclonal seeding in colorectal cancer metastasis. Ann Oncol 28(9): 2135-2141. 10.1093/annonc/mdx278
- Hur K, Toiyama Y, Schetter AJ, Okugawa Y, Harris CC, Boland CR, Goel A (2015). Identification of a metastasis-specific microRNA signature in human colorectal cancer. J Natl Cancer Inst 107(3): dju492. 10.1093/jnci/dju492
- Brabletz T, Kalluri R, Nieto MA, Weinberg RA (2018). EMT in cancer. Nat Rev Cancer 18(2): 128-134. 10.1038/nrc.2017.118
- Yook JI, Li XY, Ota I, Fearon ER, Weiss SJ (2005). Wnt-dependent regulation ft he E-cadherin repressor snail. J Biol Chem 280(12): 11740-11748. 10.1074/jbc.M413878200
- Chaw SY, Abdul Majeed A, Dalley AJ, Chan A, Stein S, Farah CS (2012). Epithelial to mesenchymal transition (EMT) biomarkers—E-cadherin, beta-catenin, APC and Vimentin—in oral squamous cell carcinogenesis and transformation. Oral Oncol 48(10): 997-1006. 10.1016/j.oraloncology.2012.05.011
- Wang H, Shi J, Luo Y, Liao Q, Niu Y, Zhang F, Shao Z, Ding Y, Zhao L (2014). LIM and SH3 protein 1 induces TGF beta-mediated epithelial-mesenchymal transition in human colorectal cancer by regulating S100A4 expression. Clin Cancer Res 20(22): 5835-5847. 10.1158/1078-0432.CCR-14-0485
- Lu Z, Ghosh S, Wang Z, Hunter T (2003). Downregulation of cavcolin-1 function by EGF leads ot he loss of E-cadherin, increased transcriptional activity of beta-catenin, and enhanced tumor cell invasion. Cancer Cell 4(6): 499-515. 10.1016/S1535-6108(03)00304-0
- Rai A, Greening DW, Chen M, Xu R, Ji H, Simpson RJ (2019). Exosomes derived from human primary and metastatic colorectal cancer cells contribute to functional heterogeneity of activated fibroblasts by reprogramming their proteome. Proteomics 19(8): e1800148. 10.1002/pmic.201800148
- Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, Wang Y, Halder G, Finegold MJ, Lee JS, Johnson RL (2010). Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci U S A 107(4): 1437-1442. 10.1073/pnas.0911427107
- Kim W, Khan SK, Gvozdenovic-Jeremic J, Kim Y, Dahlman J, Kim H, Park O, Ishitani T, Jho EH, Gao B, Yang Y (2017). Hippo signaling interactions with Wnt/beta-catenin and Notch signaling repress liver tumorigenesis. J Clin Invest 127: 137-152. 10.1172/JCI88486
- Yu FX, Zhao B, Guan KL (2015). Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163(4): 811-828. 10.1016/j.cell.2015.10.044
- Pitteri SJ, Kelly-Spratt KS, Gurley KE, Kennedy J, Buson TB, Chin A, Wang H, Zhang Q, Wong CH, Chodosh LA, Nelson PS, Hanash SM, Kemp CJ (2011). Tumor microenvironment-derived proteins dominate the plasma proteome response during breast cancer induction and progression. Cancer Res 71(15): 5090-5100. 10.1158/0008-5472.CAN-11-0568
- Pichot CS, Arvanitis C, Hartig SM, Jensen SA, Bechill J, Marzouk S, Yu J, Frost JA, Corey SJ (2010). Cdc42-interacting protein 4 promotes breast cancer cell invasion and formation of invadopodia through activation of N-WASp. Cancer Res 70(21): 8347-8356. 10.1158/0008-5472.CAN-09-4149
- Johnston AL, Lun X, Rahn JJ, Liacini A, Wang L, Hamilton MG, Parney IF, Hempstead BL, Robbins SM, Forsyth PA, Senger DL (2007). The p75 neurotrophin receptor is a central regulator of glioma invasion. PLoS Biol 5(8): e212. 10.1371/journal.pbio.0050212
- Wang L, Rahn JJ, Lun X, Sun B, Kelly JJ, Weiss S, Robbins SM, Forsyth PA, Senger DL (2008). Gamma-secretase represents a therapeutic target ort he treatment of invasive glioma mediated by the p75 neurotrophin receptor. PloS Biol 6(11): e289. 10.1371/journal.pbio.0060289
- Jabbour E, Kantarjian H (2018). Chronic myeloid leukemia: 2018 update on diagnosis, therapy and monitoring. Am J Hematol 93(3): 442-459. 10.1002/ajh.25011
- Apperley JF (2015). Chronic myeloid leukaemia. Lancet 385(9976): 1447-1459. 10.1016/S0140-6736(13)62120-0
- Peng Y, Croce CM (2016). The role of MicroRNAs in human cancer. Signal Transduct Target Ther 1: 15004. 10.1038/sigtrans.2015.4
- Yeh CH, Moles R, Nicot C (2016). Clinical significance of microRNAs in chronic and acute human leukemia. Mol Cancer 15(1): 37. 10.1186/s12943-016-0518-2
- Zhao H, Wang D, Du W, Gu D, Yang R (2010). MicroRNA and leukemia: tiny molecule, great function. Crit Rev Oncol Hematol 74(3): 149-155. 10.1016/j.critrevonc.2009.05.001
–
ACKNOWLEDGMENTS
This work was supported by National Natural Science Foundation of China (81900156 and 81821002), Science and Technology Program of Sichuan Province (2019JDTD0013), China Postdoctoral Science Foundation (2018M640927), Post-Doctor Research Pro-ject, West China Hospital, Sichuan University (2018HXBH022), and the 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (ZYJC18030 and ZYGD20008).
COPYRIGHT
© 2021

PDLIM1: Structure, function and implication in cancer by Zhou et al. is licensed under a Creative Commons Attribution 4.0 International License.