Back to article: Genotoxic stress signalling as a driver of macrophage diversity
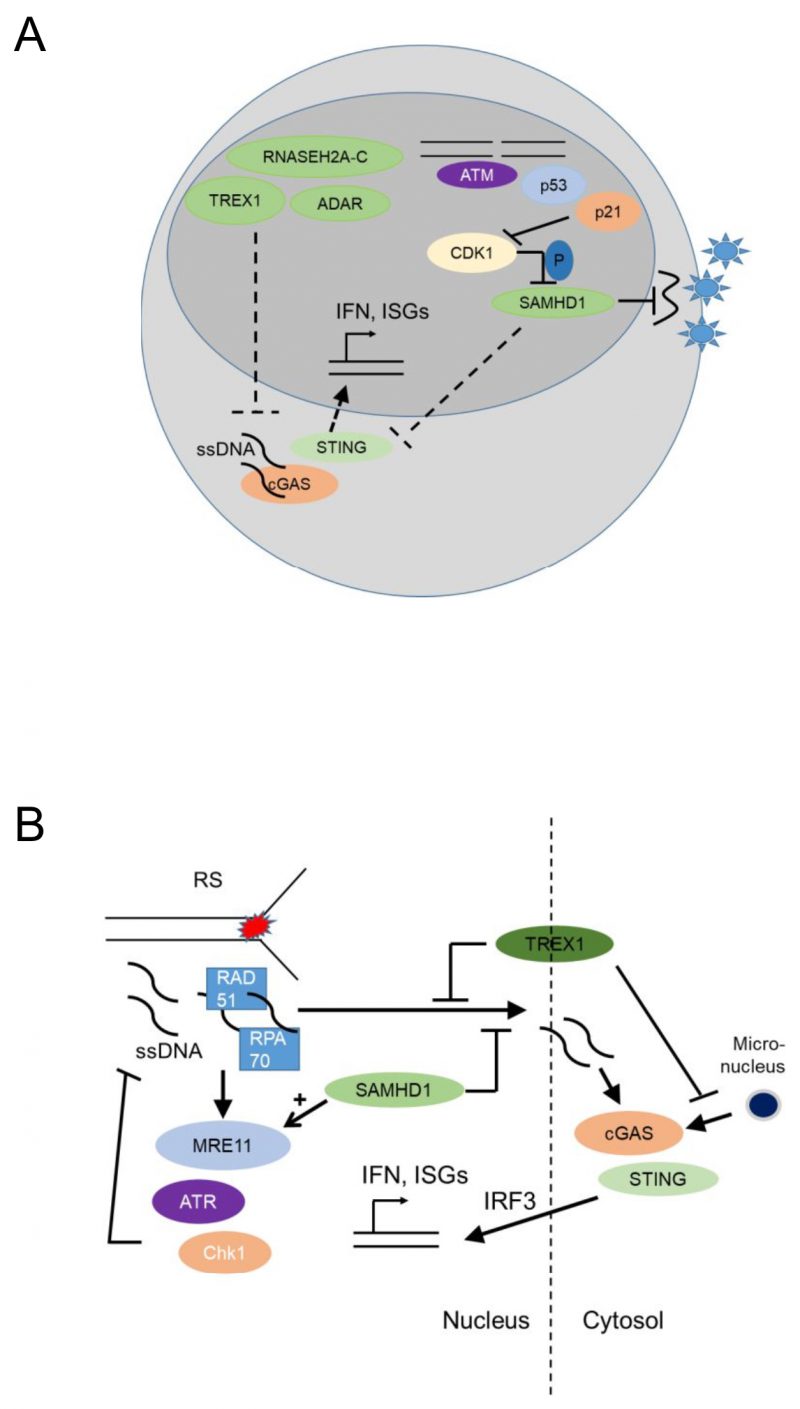
FIGURE 4: Nucleic acid metabolic pathways control antiviral programs and autoinflammation and may have genome-protecting functions by suppressing cytoplasmic cGAS activation and type I interferon responses. (A) Nucleic acid-metabolizing enzymes SAMHD1 (SAM domain and HD domain-containing protein 1), TREX1 (three prime repair exonuclease 1), RNASEH2A-C (Ribonuclease H2, Subunit A, C-Term), and ADAR (Adenosine Deaminase RNA Specific), control systemic type I interferon (IFN) levels by negatively regulating the cGAS-STING pathway. Unphosphorylated SAMHD1 restricts viral replication in myeloid cells either by depleting the dNTP pool or through its RNAse activity. This antiviral activity of SAMHD1 is suppressed upon SAMHD1 phosphorylation by cyclin A2/Cdk1 in cycling cells, while genotoxic stress that leads to p53 and p21 upregulation and subsequently CDK1/2 (cyclin-dependent kinase 1/2) suppression and SAMHD1 dephosphorylation increase this antiviral activity. (B) SAMHD1 promotes degradation of ssDNA at stalled replication forks by stimulating the exonuclease activity of MRE11 and thus activating ATR-CHK1 to alleviate replication stress. This prevents ssDNA accumulation in the cytosol where they would activate cGAS-STING leading to increased expression of type I interferons. TREX1 (three prime repair exonuclease 1) also promotes the removal of ssDNA molecules (formed due to replication stress) through RPA70 (Replication protein A 70 kDa DNA-binding subunit) and Rad51, thus preventing leakage of ssDNA to the cytosol and phosphorylation of IRF3. TREX1 also limits cGAS activation at micronuclei, by degrading micronuclear DNA. Replication stress, ssDNA and micronuclei promote the cGAS – STING – type I IFN activation pathway.