Back to article: Assay for high-throughput screening of inhibitors of the ASC-PYD inflammasome core filament
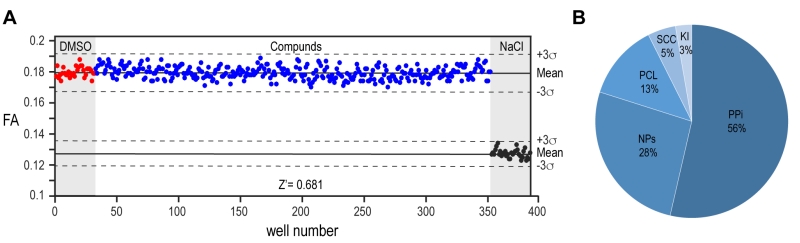
FIGURE 4: High-throughput screening of compound library. (A) Evaluation and visualization of fluorescence anisotropy data from the high-throughput screening of 10,100 compounds. The screening is performed on 384-well plates where the first and the last two columns are used for negative controls reported in red (0.1% DMSO; no drug added) and positive controls reported in black (900 mM NaCl). The areas used for positive and negative controls are highlighted in light grey. The middle columns (320 wells) are used to test compounds (10 μM compound final concentration, final DMSO concentration 0.1%) and reported in blue. The library was processed in duplicate on 64 plates. The Z’-factor determination for plate validation is calculated using the fluorescence anisotropy signals of the controls and compounds wells. The solid lines represent the mean fluorescence anisotropy value for controls and compounds measurements, the dashed lines denote the average ± 3 standard deviation of the negative and the positive controls, respectively. In this example, a Z’-factor of 0.681 is suitable to validate the plate. (B) Pie chart of 10,100 compounds from BSF-ACCESS Chemical Collection. Five different classes of compounds are screened: PPi (Protein-Protein interaction, 5’411 compounds), NPs (Natural Products, 2’654 compounds), PCL (The Prestwick Chemical Library, 1’280 compounds), SCC (Swiss Chemical Collection, 474 compounds) and Kinase Inhibitors, 275 compounds. The relative percentage of each class of compounds over the entire screened library is reported.