News and thoughts:
Cell Stress, Vol. 1, No. 1, pp. 7 - 10; doi: 10.15698/cst2017.10.103
TFEB-mTORC1 feedback loop in metabolism and cancer
1 Telethon Institute of Genetics and Medicine (TIGEM), via Campi Flegrei, 34, 80078 Pozzuoli (Naples), Italy.
2 Jan and Dan Duncan Neurological Research Institute, Houston, TX 77030, USA.
3 Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas 77030, USA.
4 Medical Genetics Unit, Department of Medical and Translational Science, Federico II University, Via Pansini 5, 80131 Naples, Italy.
Keywords: mTORC1, TFEB, RAGD, transcription, metabolism, cancer.
Received originally: 20/09/2017 Accepted: 25/09/2017
Published: 01/10/2017
Correspondence:
Andrea Ballabio, Telethon Institute of Genetics and Medicine (TIGEM), via Campi Flegrei, 34, 80078 Pozzuoli (Naples), Italy ballabio@tigem.it
Conflict of interest statement: There is no conflict of interest regarding the publication of this article.
Please cite this article as: Chiara Di Malta and Andrea Ballabio (2017). TFEB-mTORC1 feedback loop in metabolism and cancer Cell Stress 1(1): 7-10. doi: 10.15698/cst2017.10.103
Everyday most organisms face the requirement to adapt their metabolic cues to the fluctuation of nutrient resources into the environment. When nutrients are abundant cells activate anabolic pathways, such as protein and lipid synthesis, and promote cell proliferation and growth. On the contrary, when food availability is scarce or even absent, cells shut off anabolism and activate a catabolic program that guarantees their own survival through the generation of energy from the degradation of intracellular substrates [1].
–
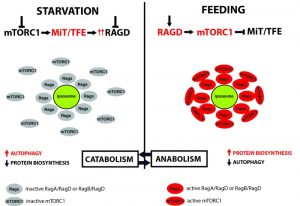
A key regulator of metabolic adaptation to nutrient availability is the mTOR Complex 1 (mTORC1) kinase, which promotes anabolic and limits catabolic pathways through the phosphorylation of target substrates [2]. Multiple inputs modulate mTORC1 activity, in particular amino acids are essential for its full activation. Amino acids mediate mTORC1 activation through the highly-conserved family of RagGTPases. Mammals have four types of Rag GTPases, Rag A, B, C and D, which form RagA or RagB/RagC or RagD obligate heterodimers [3]. In presence of amino acids Rag proteins are activated through a GDP/GTP molecular switch that enables them to interact with the raptor subunit of mTORC1 to mediate its re-localization from the cytoplasm to the lysosome [4][5][6]. mTORC1 lysosomal localization is a “sine qua non” condition for its activation, since its activator Rheb also localizes to the lysosome [7].
–
MiT/TFE transcription factors (TFs) are master regulators of lysosomal and melanosomal biogenesis and autophagy and their activity is inhibited by mTORC1 [8][9][10][11][12][13]. In presence of nutrients mTORC1 phosphorylates MiT/TFE TFs at critical serine residues, leading to their cytoplasmic retention. RagGTPases are also directly implicated in this event, since active Rags interact with MiT/TFE TFs and contribute to their lysosomal recruitment [14].
–
During starvation, inhibition of mTORC1 and activation of the phosphatase calcineurin lead to MiT/TFE de-phosphorylation and nuclear translocation [15]. Once in the nucleus, they induce expression of a large group of genes that support cellular catabolism, such as lysosomal and autophagy genes. In this way, the cell regulates the expression of catabolic genes according to nutrient availability.
–
In our recent work we postulated that MiT/TFE TFs, which are regulated by mTORC1, could in turn influence mTORC1 activity. To test this hypothesis we analyzed mTORC1 signaling in cells and in tissues of mice in which the activity of MiT/TFE TFs was artificially manipulated.
–
We observed that overexpression of TFEB or TFE3 increased mTORC1 activation whereas their depletion significantly impaired mTORC1 signaling upon nutrient stimulation both in vitro and in vivo. Thus, the activation of mTORC1 in response to nutrients is regulated by MiT/TFE TFs.
–
mTORC1 promotes protein synthesis in muscles upon physical exercise to support muscle growth [16]. This effect is well appreciated by bodybuilders, who know that eating a protein-rich meal after exercise is a good strategy to increase muscle mass. However the mechanisms underlying this process are still poorly understood. Interestingly, TFEB nuclear translocation is induced upon physical exercise as consequence of calcineurin phosphatase activation [15]. Therefore, we hypothesized that TFEB could boost mTORC1 signaling in response to a meal after exercise. Consistently, exercised muscle-specific TFEB KO mice showed a reduced induction of mTORC1 activity and protein synthesis in response to leucine after exercise, indicating that mTORC1 activation upon physical exercise requires MiT/TFE TFs. Our findings suggested the existence of a new, physiologically relevant, molecular pathway, that regulates mTORC1 activity through MiT/TFE TFs.
–
Next, we investigated the mechanism underlying this regulation. We tested the hypothesis that MiT/TFE genes regulate the expression of genes that are important for mTORC1 activity. We searched for the presence of multiple MiT/TFE DNA binding sites, defined as “CLEAR elements”, in the proximal promoters of 50 human genes encoding mTORC1-related genes.
–
Among the putative candidates, the RagD GTPase mRNA levels were the most significantly increased as a consequence of TFEB overexpression. RagC transcript levels, as well as those of Folliculin (FLCN), a GTPase activating protein (GAP) for RagC/D [17], were also increased, albeit at lower levels, whereas RagB and RagA were unchanged. Opposite effects were observed by TFEB or TFE3 siRNA-mediated silencing. Chromatin immuno-Precipitation (ChiP) and luciferase assay demonstrated that RagD is a direct target of TFEB. Importantly, CRISPR-mediated deletion of the MiT/TFE response element in the RagD promoter suppressed RagD expression and mTORC1 activation upon amino acid stimulation. In line with the notion that Rag GTPases mediates mTORC1 lysosomal localization, we found that mTOR localization on the lysosome correlated with TFEB levels, being significantly decreased in cells lacking the RagD promoter region bound by MiT/TFE factors.
–
How modulation of RagD levels alone can be sufficient to significantly impair mTORC1 signaling remains an open question. One would expect that RagC could compensate for RagD but our study suggest that these two Rags are less interchangeable than what was previously thought. Another critical point is that to be functionally active RagD needs to form heterodimers with either RagA or B. We noticed that RagD levels are lower compared to the other Rag GTPases (data not shown), therefore, one possibility is that RagD represents a limiting factor for RagD/RagA and RagD/RagB heterodimer formation. In line with this hypothesis is the observation that perturbing MiT/TFE mediated transcriptional regulation of RagD only is sufficient to severely hampers mTORC1 lysosomal recruitment and hence its activation.
–
In summary, we defined a new regulation of mTORC1 activity in response to nutrients. Starvation inhibits mTORC1 thus allowing MiT/TFE nuclear translocation and hence activation of catabolism via the expression of autophagic and lysosomal genes. At the same time, MiT/TFE TFs induce the expression of RagD and this results in the assembly of inactive Rags heterodimers on the lysosomal surface. Feeding turns on Rags which can now recruit mTORC1 to the lysosome and promote its activation. In this way, the cell gets ready to efficiently switch on anabolism and turn off catabolism when nutrients become available (Fig. 1).
–
–
Constitutive activation of MiT/TFE TFs drives tumorigenesis in several cancer types, such as Renal Cell Carcinoma (RCC), melanoma and pancreatic ductal adenocarcinoma (PDA) [18][19][20]. However, the role of MiT/TFE factors in oncogenesis is still unclear. We demonstrated that constitutive induction of RagD GTPase and mTORC1 hyper-activation are hallmarks of MiT/TFE-dependent RCC, PDA and melanoma. Importantly, we found that silencing of RagD strongly reduced xenograft tumor growth of melanoma cells presenting elevated RagD expression and increased mTORC1 activation, indicating that RagD contributes to tumor cell growth in these malignancies. These findings define a new oncogenic pathway in MiT/TFE dependent tumors with potential implications for therapy.
–
Interestingly, clinical data indicated that patients affected by RCC caused by TFE3 chromosomal translocation frequently presented activation of mTOR signaling with variable suppression of cancer growth by treatment with mTOR inhibitors [18]. Our study provides the molecular mechanism for these clinical observations and encourages further research on the potentiality of this therapeutic approach for MiT/TFE dependent malignancies.
References
- J.D. Rabinowitz, and E. White, "Autophagy and Metabolism", Science, vol. 330, pp. 1344-1348, 2010. http://dx.doi.org/10.1126/science.1193497
- R.A. Saxton, and D.M. Sabatini, "mTOR Signaling in Growth, Metabolism, and Disease", Cell, vol. 169, pp. 361-371, 2017. http://dx.doi.org/10.1016/j.cell.2017.03.035
- T. Sekiguchi, E. Hirose, N. Nakashima, M. Ii, and T. Nishimoto, "Novel G Proteins, Rag C and Rag D, Interact with GTP-binding Proteins, Rag A and Rag B", Journal of Biological Chemistry, vol. 276, pp. 7246-7257, 2001. http://dx.doi.org/10.1074/jbc.M004389200
- Y. Sancak, T.R. Peterson, Y.D. Shaul, R.A. Lindquist, C.C. Thoreen, L. Bar-Peled, and D.M. Sabatini, "The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to mTORC1", Science, vol. 320, pp. 1496-1501, 2008. http://dx.doi.org/10.1126/science.1157535
- E. Kim, P. Goraksha-Hicks, L. Li, T.P. Neufeld, and K. Guan, "Regulation of TORC1 by Rag GTPases in nutrient response", Nature Cell Biology, vol. 10, pp. 935-945, 2008. http://dx.doi.org/10.1038/ncb1753
- Y. Sancak, L. Bar-Peled, R. Zoncu, A.L. Markhard, S. Nada, and D.M. Sabatini, "Ragulator-Rag Complex Targets mTORC1 to the Lysosomal Surface and Is Necessary for Its Activation by Amino Acids", Cell, vol. 141, pp. 290-303, 2010. http://dx.doi.org/10.1016/j.cell.2010.02.024
- S. Menon, C. Dibble, G. Talbott, G. Hoxhaj, A. Valvezan, H. Takahashi, L. Cantley, and B. Manning, "Spatial Control of the TSC Complex Integrates Insulin and Nutrient Regulation of mTORC1 at the Lysosome", Cell, vol. 156, pp. 771-785, 2014. http://dx.doi.org/10.1016/j.cell.2013.11.049
- M. Sardiello, M. Palmieri, A. di Ronza, D.L. Medina, M. Valenza, V.A. Gennarino, C. Di Malta, F. Donaudy, V. Embrione, R.S. Polishchuk, S. Banfi, G. Parenti, E. Cattaneo, and A. Ballabio, "A Gene Network Regulating Lysosomal Biogenesis and Function", Science, vol. 325, pp. 473-477, 2009. http://dx.doi.org/10.1126/science.1174447
- C. Settembre, C. Di Malta, V.A. Polito, M.G. Arencibia, F. Vetrini, S. Erdin, S.U. Erdin, T. Huynh, D. Medina, P. Colella, M. Sardiello, D.C. Rubinsztein, and A. Ballabio, "TFEB Links Autophagy to Lysosomal Biogenesis", Science, vol. 332, pp. 1429-1433, 2011. http://dx.doi.org/10.1126/science.1204592
- J.A. Martina, H.I. Diab, L. Lishu, L. Jeong-A, S. Patange, N. Raben, and R. Puertollano, "The Nutrient-Responsive Transcription Factor TFE3 Promotes Autophagy, Lysosomal Biogenesis, and Clearance of Cellular Debris", Science Signaling, vol. 7, 2014. http://dx.doi.org/10.1126/scisignal.2004754
- E. Steingrímsson, L. Tessarollo, B. Pathak, L. Hou, H. Arnheiter, N.G. Copeland, and N.A. Jenkins, "Mitf and Tfe3, two members of the Mitf-Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development", Proceedings of the National Academy of Sciences, vol. 99, pp. 4477-4482, 2002. http://dx.doi.org/10.1073/pnas.072071099
- C. Settembre, R. Zoncu, D.L. Medina, F. Vetrini, S. Erdin, S. Erdin, T. Huynh, M. Ferron, G. Karsenty, M.C. Vellard, V. Facchinetti, D.M. Sabatini, and A. Ballabio, "A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB", The EMBO Journal, vol. 31, pp. 1095-1108, 2012. http://dx.doi.org/10.1038/emboj.2012.32
- A. Roczniak-Ferguson, C.S. Petit, F. Froehlich, S. Qian, J. Ky, B. Angarola, T.C. Walther, and S.M. Ferguson, "The Transcription Factor TFEB Links mTORC1 Signaling to Transcriptional Control of Lysosome Homeostasis", Science Signaling, vol. 5, 2012. http://dx.doi.org/10.1126/scisignal.2002790
- J.A. Martina, and R. Puertollano, "Rag GTPases mediate amino acid–dependent recruitment of TFEB and MITF to lysosomes", Journal of Cell Biology, vol. 200, pp. 475-491, 2013. http://dx.doi.org/10.1083/jcb.201209135
- D.L. Medina, S. Di Paola, I. Peluso, A. Armani, D. De Stefani, R. Venditti, S. Montefusco, A. Scotto-Rosato, C. Prezioso, A. Forrester, C. Settembre, W. Wang, Q. Gao, H. Xu, M. Sandri, R. Rizzuto, M.A. De Matteis, and A. Ballabio, "Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB", Nature Cell Biology, vol. 17, pp. 288-299, 2015. http://dx.doi.org/10.1038/ncb3114
- K. Watson, and K. Baar, "mTOR and the health benefits of exercise", Seminars in Cell & Developmental Biology, vol. 36, pp. 130-139, 2014. http://dx.doi.org/10.1016/j.semcdb.2014.08.013
- Z. Tsun, L. Bar-Peled, L. Chantranupong, R. Zoncu, T. Wang, C. Kim, E. Spooner, and D. Sabatini, "The Folliculin Tumor Suppressor Is a GAP for the RagC/D GTPases That Signal Amino Acid Levels to mTORC1", Molecular Cell, vol. 52, pp. 495-505, 2013. http://dx.doi.org/10.1016/j.molcel.2013.09.016
- E.C. Kauffman, C.J. Ricketts, S. Rais-Bahrami, Y. Yang, M.J. Merino, D.P. Bottaro, R. Srinivasan, and W.M. Linehan, "Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers", Nature Reviews Urology, vol. 11, pp. 465-475, 2014. http://dx.doi.org/10.1038/nrurol.2014.162
- R. Haq, and D.E. Fisher, "Biology and Clinical Relevance of the Micropthalmia Family of Transcription Factors in Human Cancer", Journal of Clinical Oncology, vol. 29, pp. 3474-3482, 2011. http://dx.doi.org/10.1200/JCO.2010.32.6223
- R.M. Perera, S. Stoykova, B.N. Nicolay, K.N. Ross, J. Fitamant, M. Boukhali, J. Lengrand, V. Deshpande, M.K. Selig, C.R. Ferrone, J. Settleman, G. Stephanopoulos, N.J. Dyson, R. Zoncu, S. Ramaswamy, W. Haas, and N. Bardeesy, "Transcriptional control of autophagy–lysosome function drives pancreatic cancer metabolism", Nature, vol. 524, pp. 361-365, 2015. http://dx.doi.org/10.1038/nature14587
ACKNOWLEDGMENTS
This work was supported by grants from the Italian Telethon Foundation (TGM11CB6), the European Research Council Advanced Investigator grant no. 250154 (CLEAR) (A.B.) and no 341131 (InMec) (P.G.P.); US National Institutes of Health (R01-NS078072) (A.B.) and the Associazione Italiana per la Ricerca sul Cancro (A.I.R.C.) to A.B (IG 2015 Id 17639) and C.S. (IG 2015 Id 17717).
COPYRIGHT
© 2017

TFEB-mTORC1 feedback loop in metabolism and cancer by Di Malta and Ballabio is licensed under a Creative Commons Attribution 4.0 International License.